RNASeq transcriptome assembly folder
$ cd /data/gpfs/assoc/bch709-1/YOURID
$ mkdir rnaseq/
$ cd rnaseq
File copy
$ cd /data/gpfs/assoc/bch709-1/YOURID/
$ mkdir -p rnaseq/transcriptome_assembly/fastq
$ cd rnaseq/transcriptome_assembly/fastq
$ cp /data/gpfs/assoc/bch709-1/Course_material/2020/RNASeq/*.gz .
Conda environment installation
conda clean --all
conda env remove -n rnaseq
conda create -n rnaseq python=3
conda activate rnaseq
conda install -c conda-forge -c bioconda fastqc star rsem subread hisat2 bowtie2 samtools multiqc trim-galore trinity -y
conda install -c conda-forge/label/cf201901 nano -y
FASTQC
pwd
cd /data/gpfs/assoc/bch709-1/YOURID/rnaseq/transcriptome_assembly/fastq
fastqc pair1.fastq.gz pair2.fastq.gz
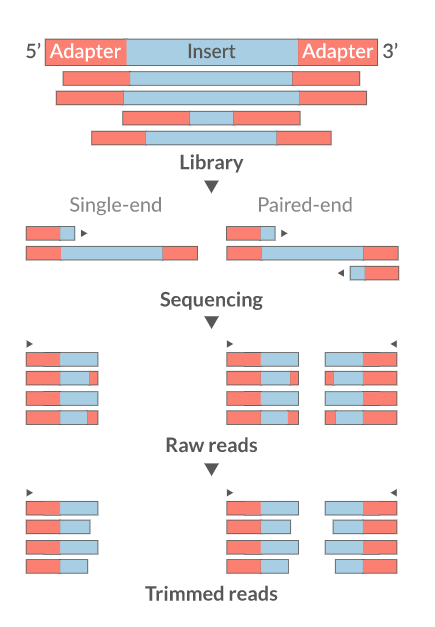
Trim the reads
- Trim IF necessary
- Synthetic bases can be an issue for SNP calling
- Insert size distribution may be more important for assemblers
- Trim/Clip/Filter reads
- Remove adapter sequences
- Trim reads by quality
- Sliding window trimming
- Filter by min/max read length
- Remove reads less than ~18nt
- Demultiplexing/Splitting
Cutadapt
fastp
Skewer
Prinseq
Trimmomatics
Trim Galore
Run trimming
trim_galore --help
trim_galore --paired --three_prime_clip_R1 5 --three_prime_clip_R2 5 --max_n 40 --gzip -o trim pair1.fastq.gz pair2.fastq.gz --core 8 --fastqc
ls trim/
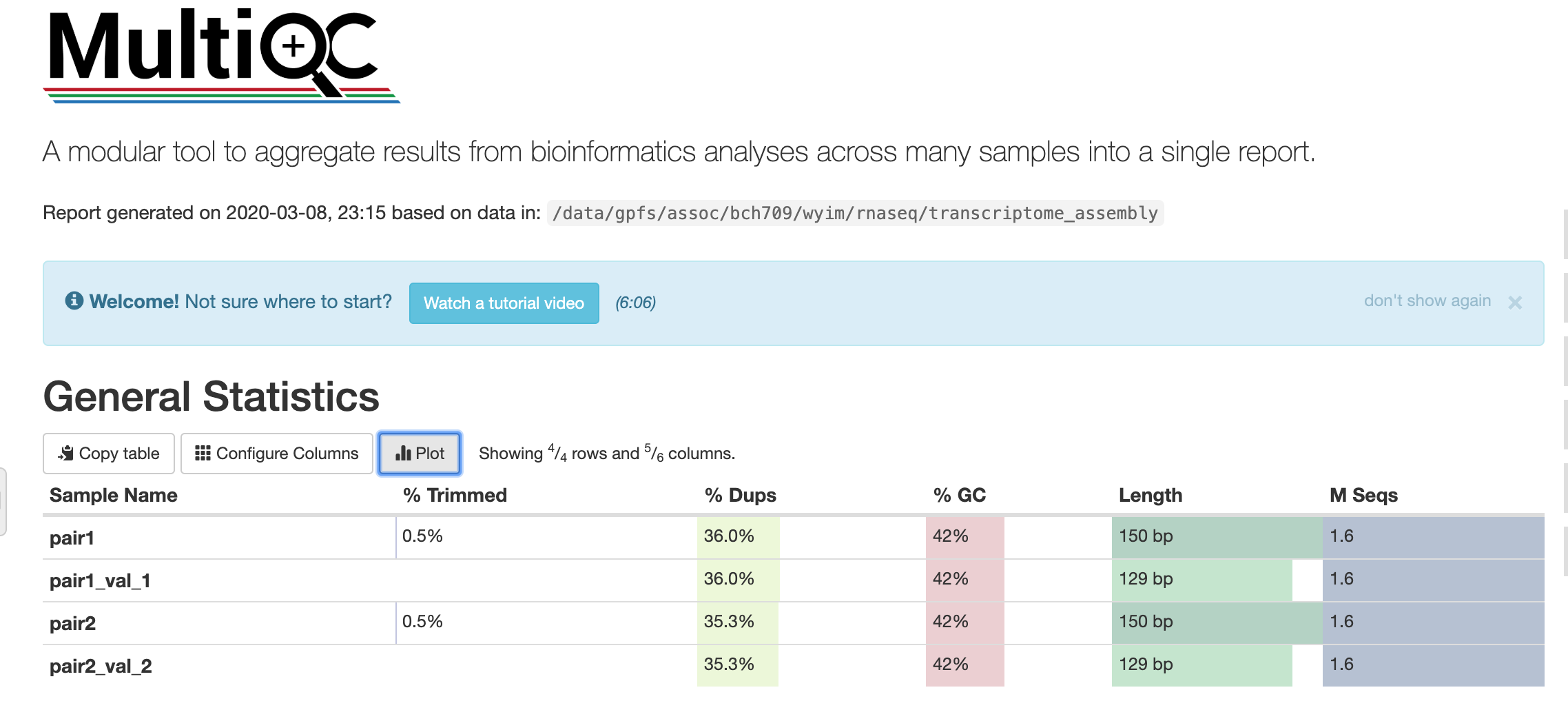
How to make a report?
multiqc --help
multiqc --filename transcriptome_assembly .
PLEASE CHECK YOUR MULTIQC with SCP from your Desktop
scp <YOURID>@pronghorn.rc.unr.edu:/data/gpfs/assoc/bch709-1/wyim/rnaseq/transcriptome_assembly/transcriptome_assembly.html ~/Desktop
Trinity assembly
Trinity --seqType fq --max_memory <MEMORY> --left <READ1> --right <READ> --CPU <CPU_NUMBER>
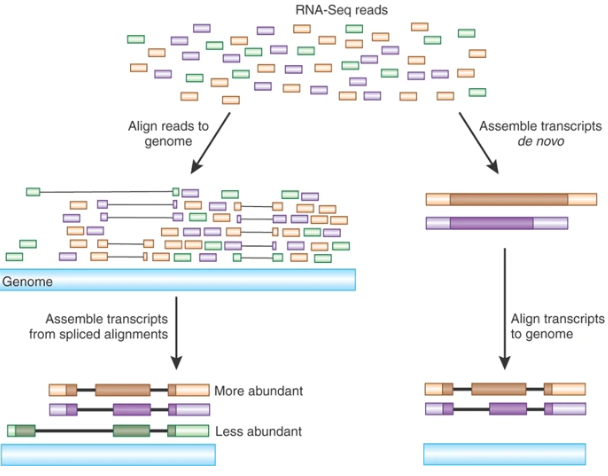
Transcriptome Assembly
De novo assembly
Assembly?


Sequencing coverage


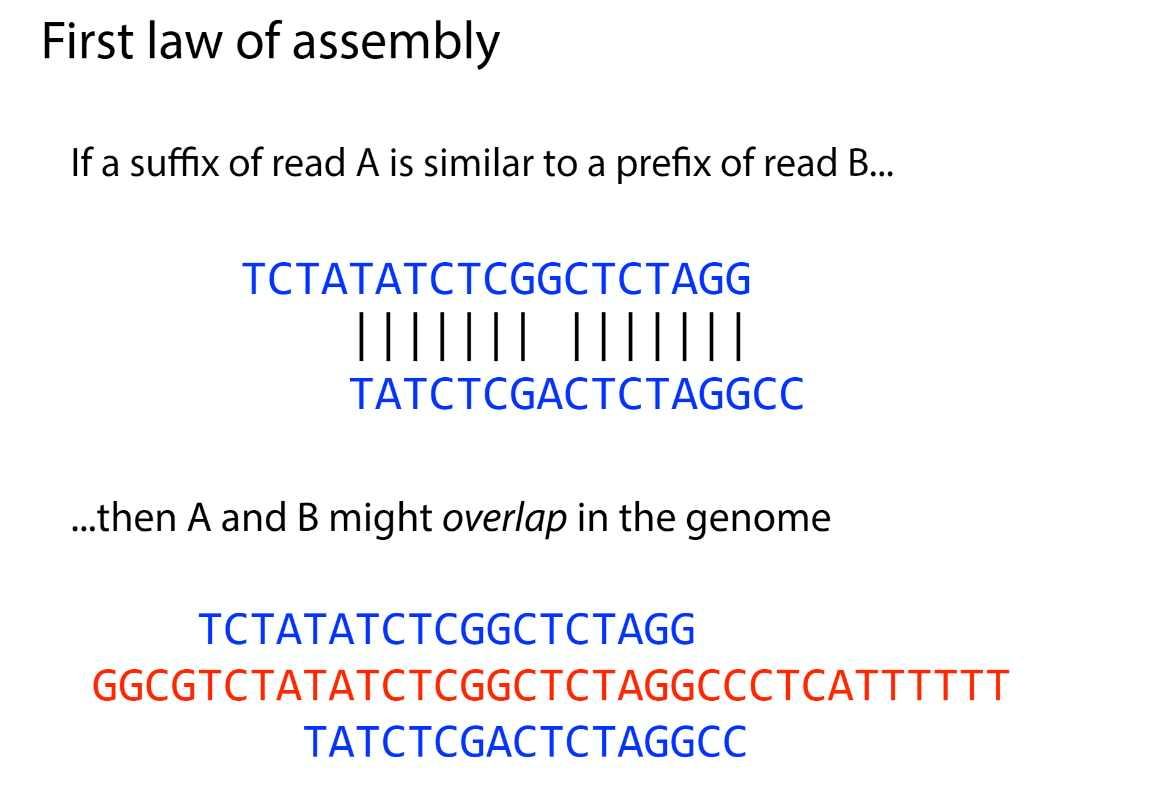
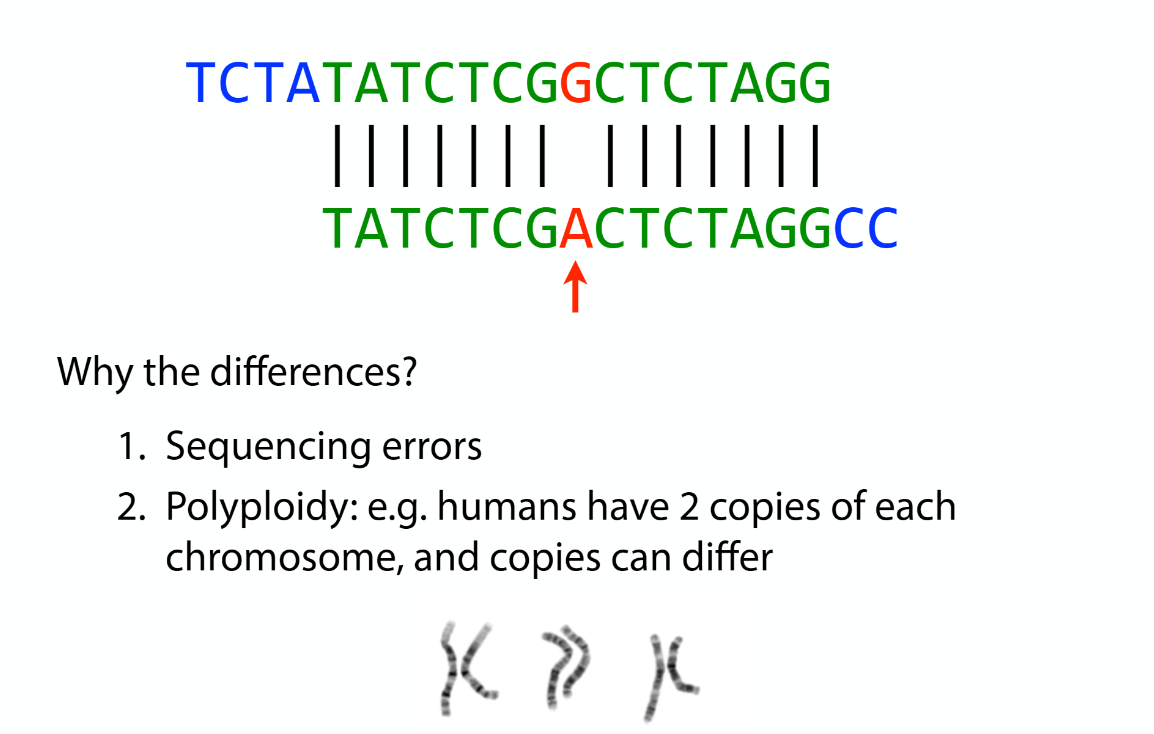
Assembly law
Overlap graph
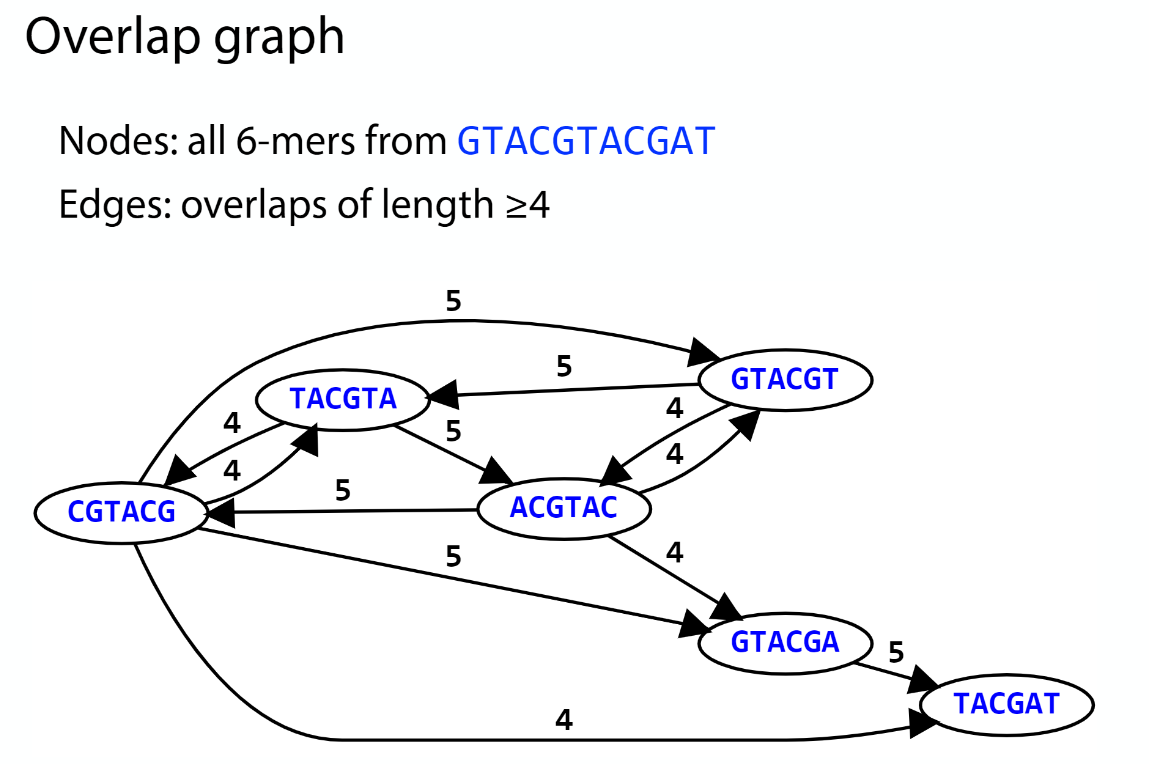
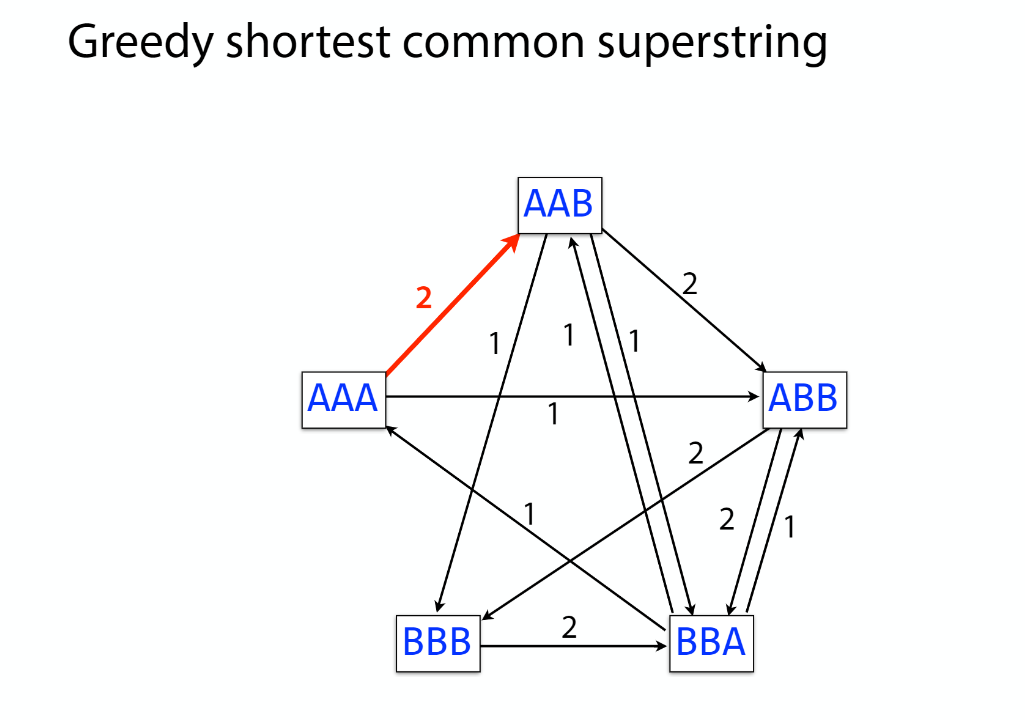
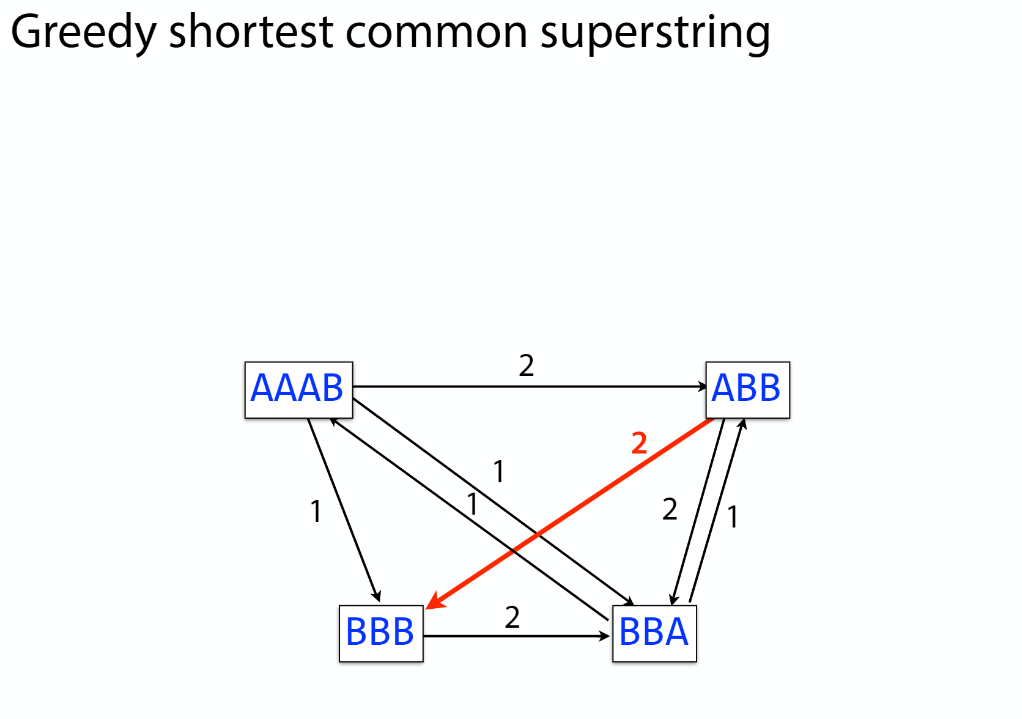
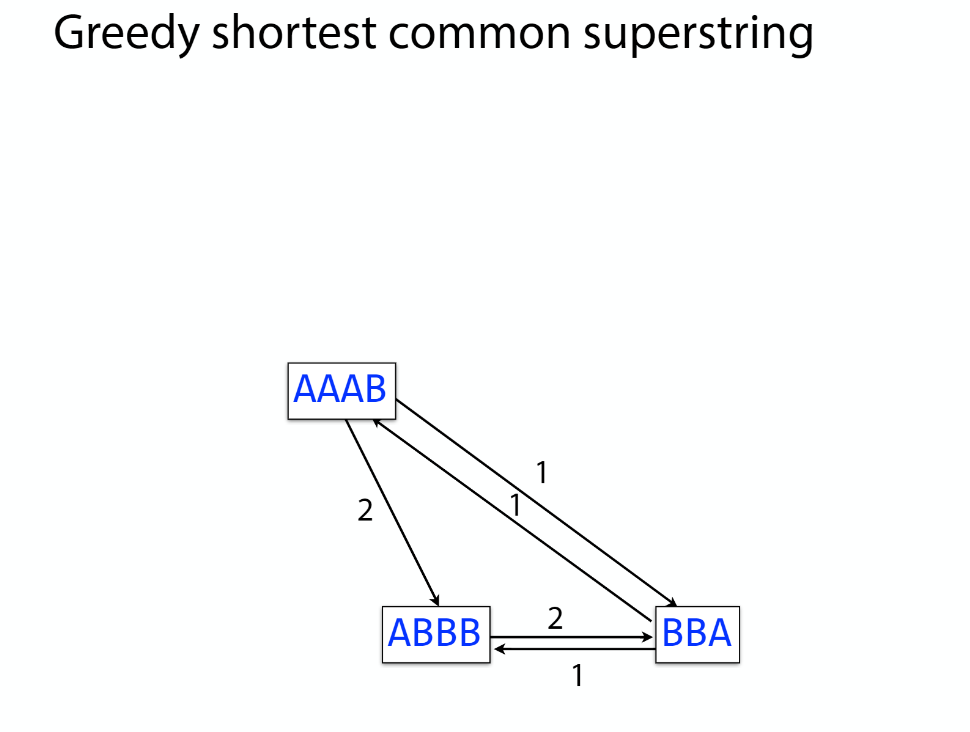
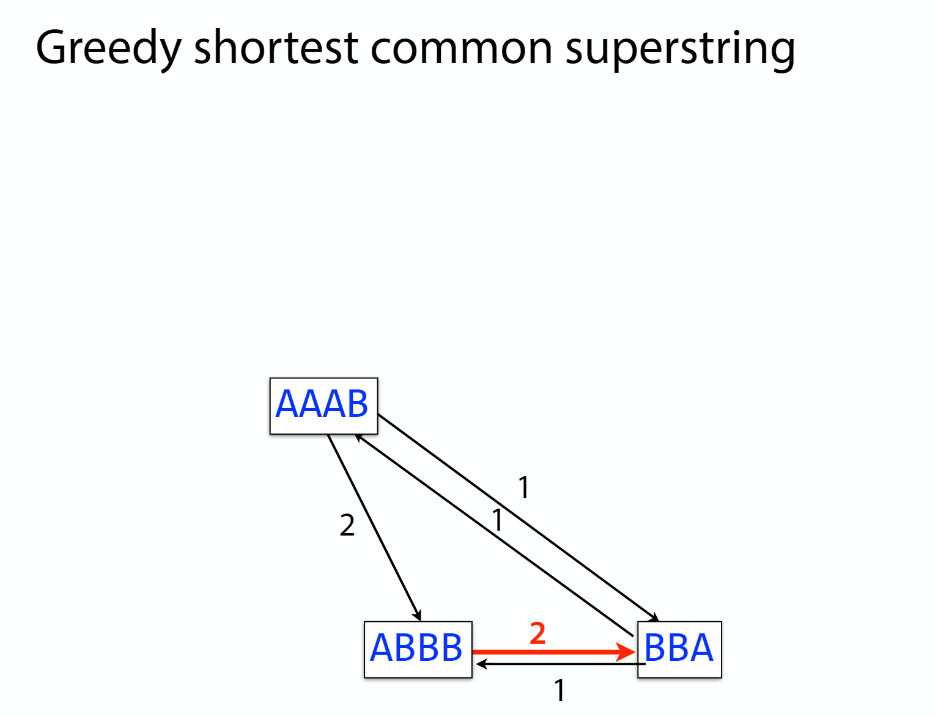
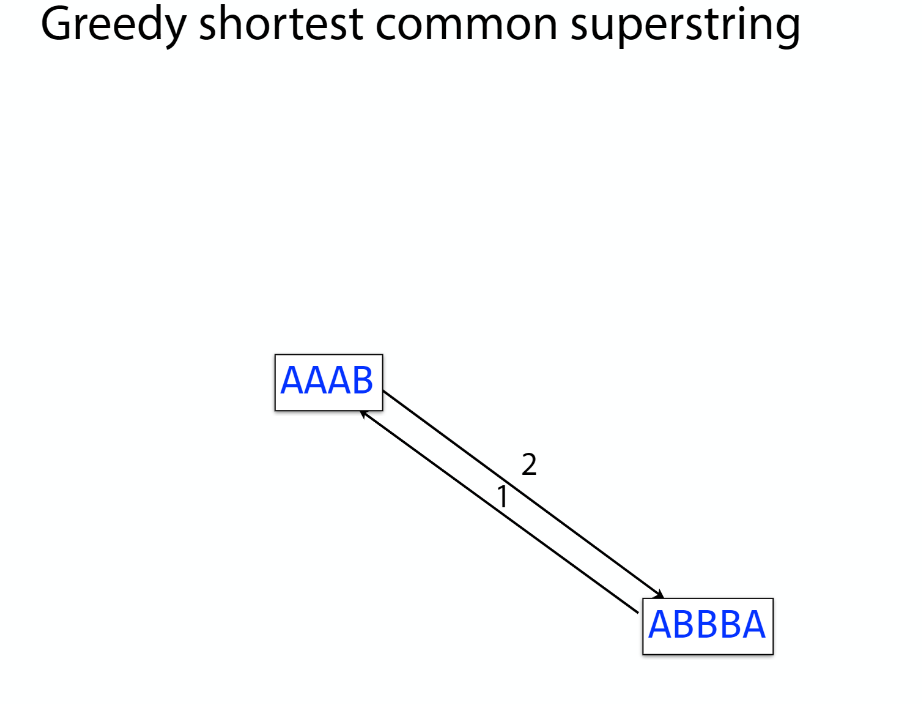
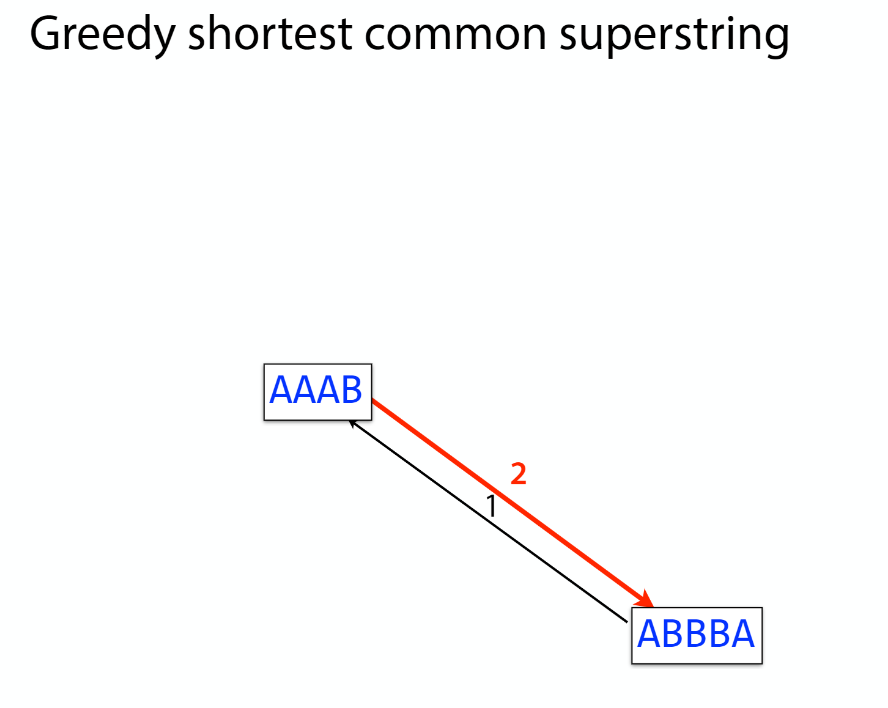
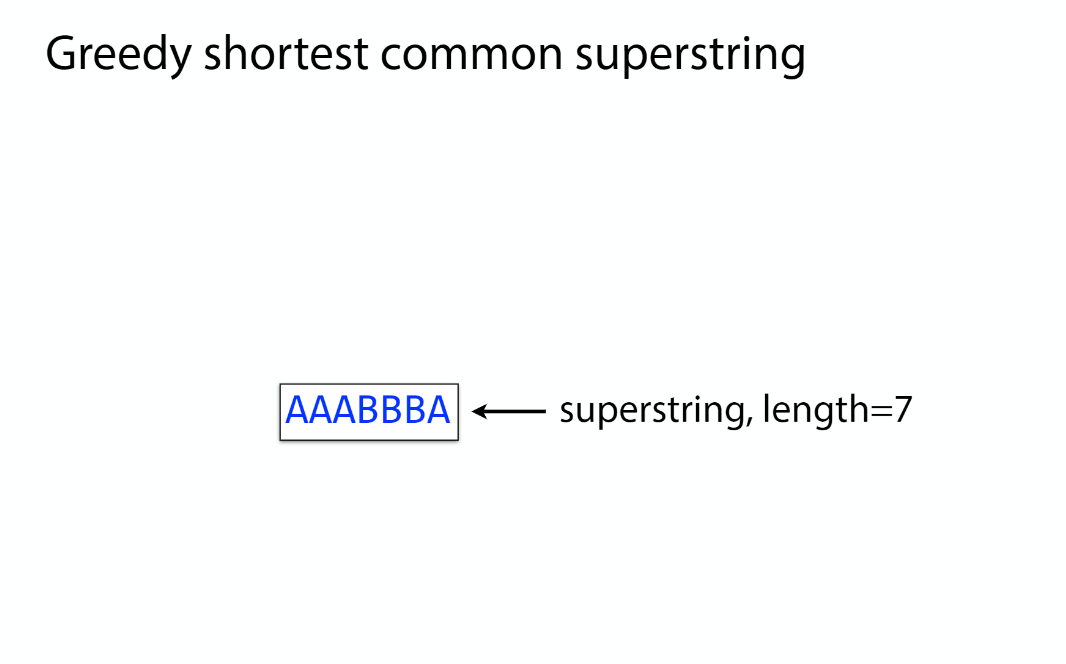

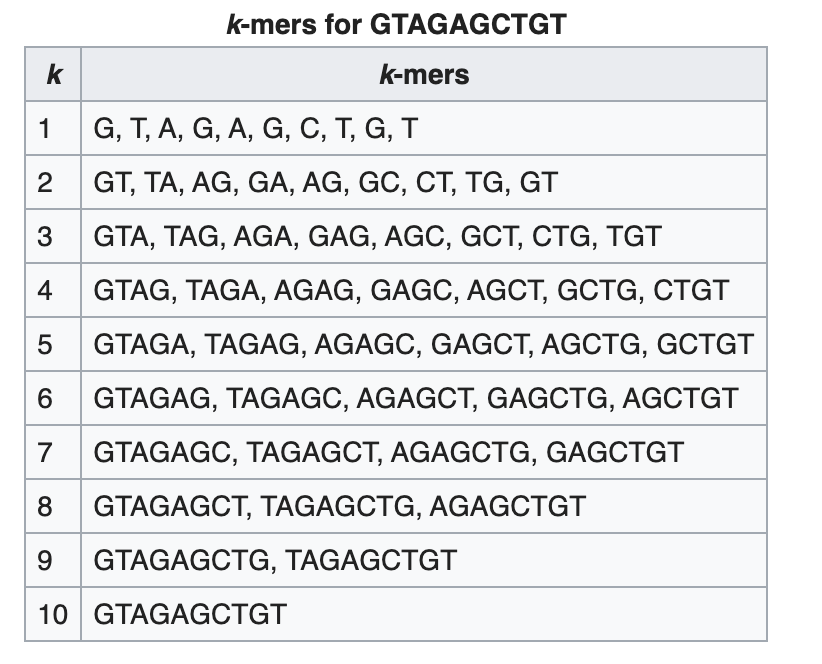
K-mer: substring of length k
k-mers are subsequences of length k contained within a biological sequence.
De bruijn
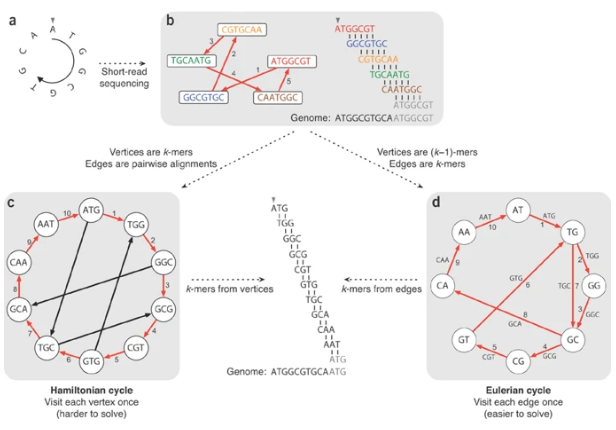
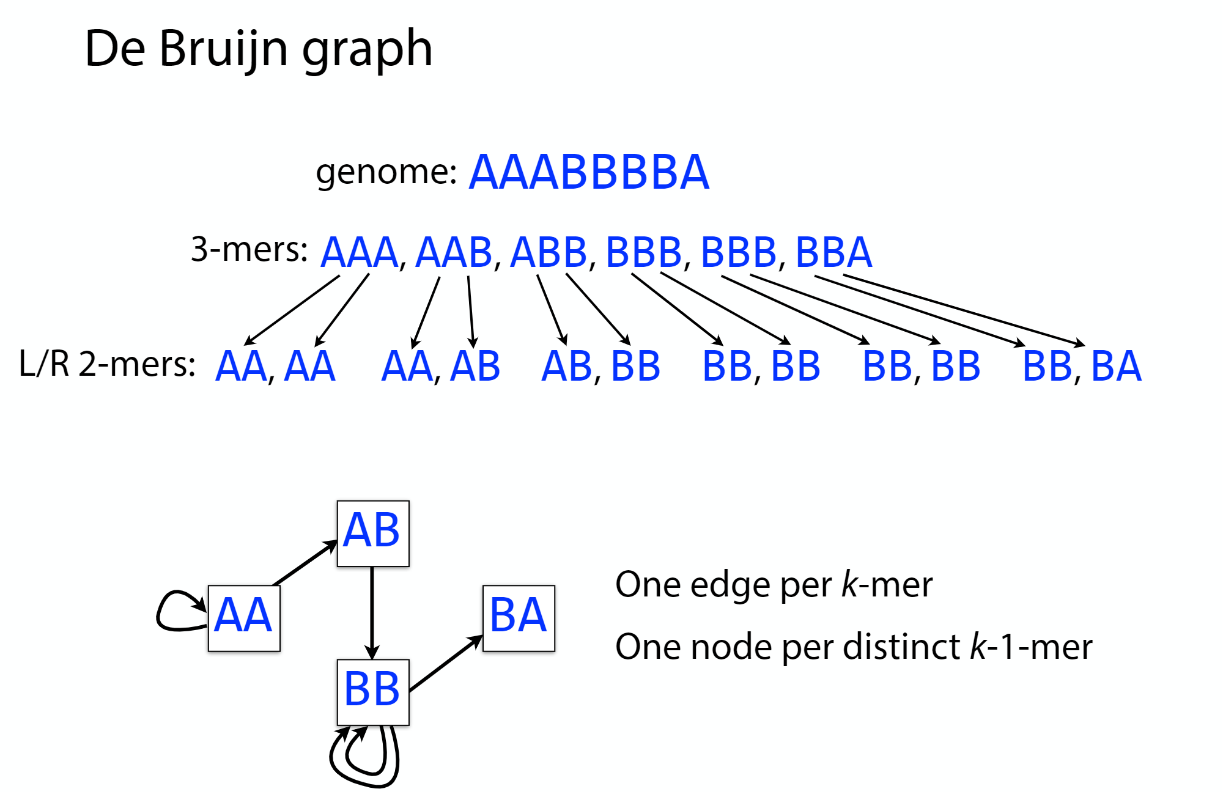
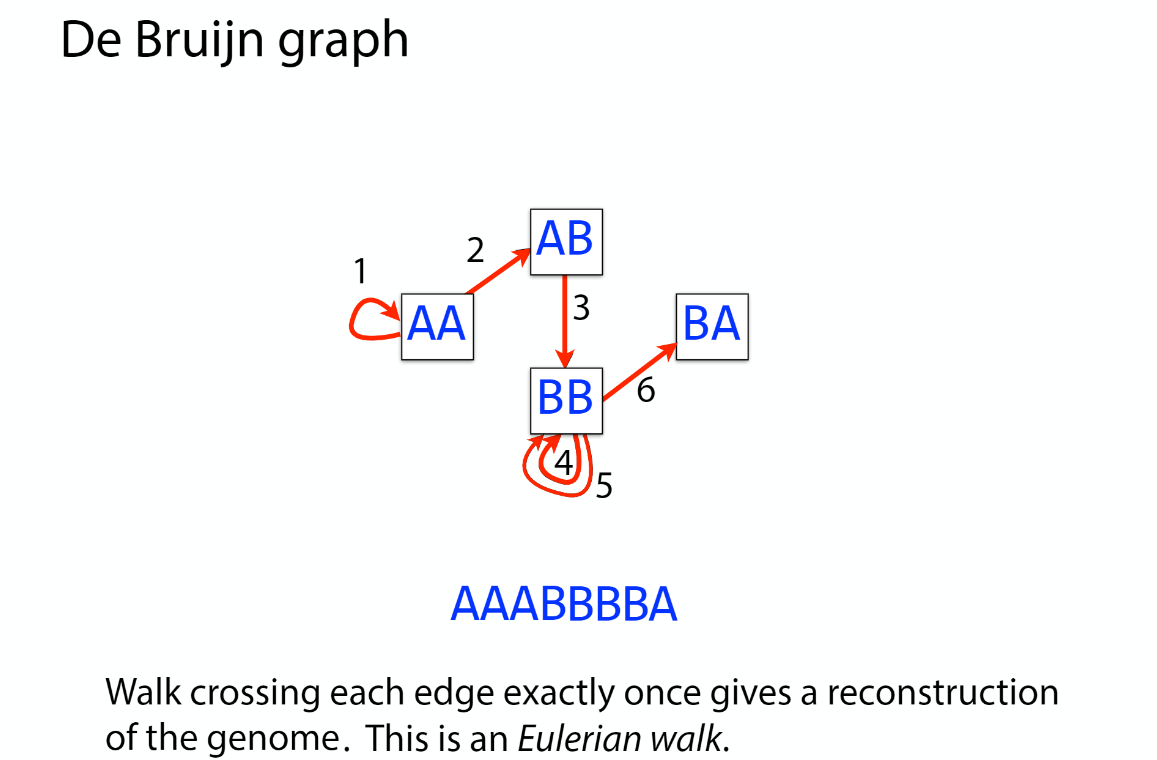
OLC vs De bruijn
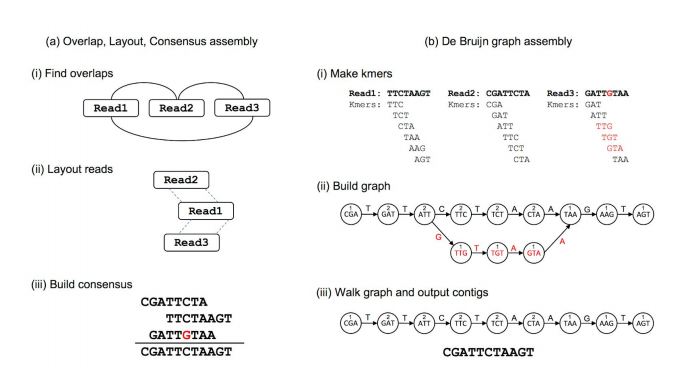
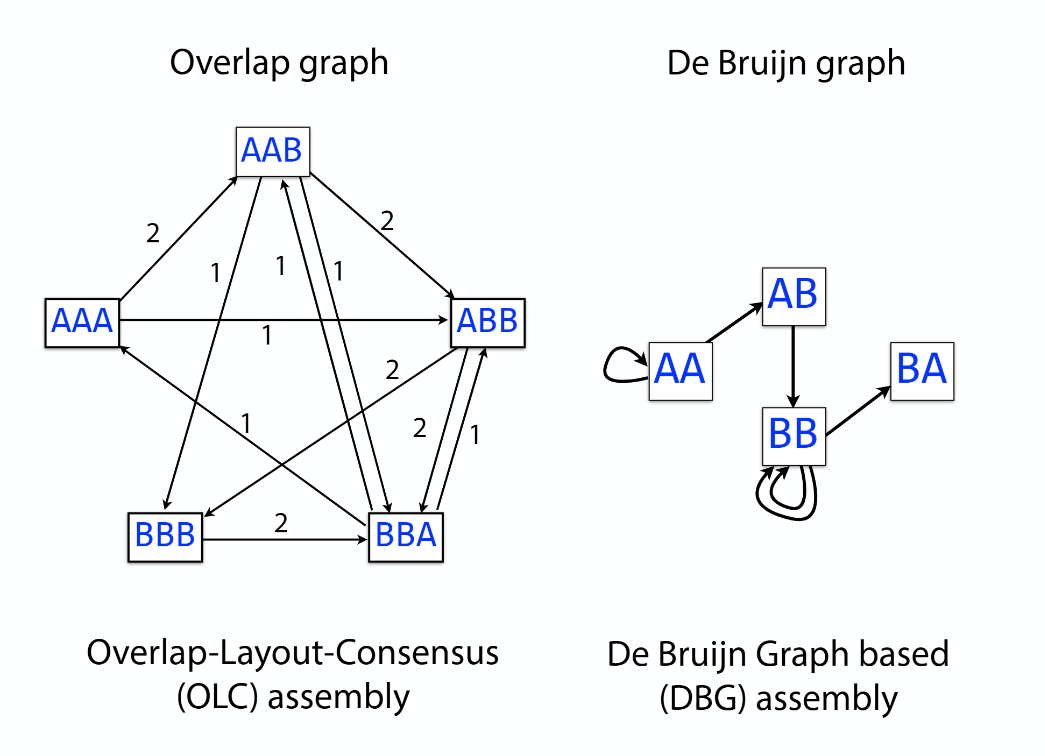
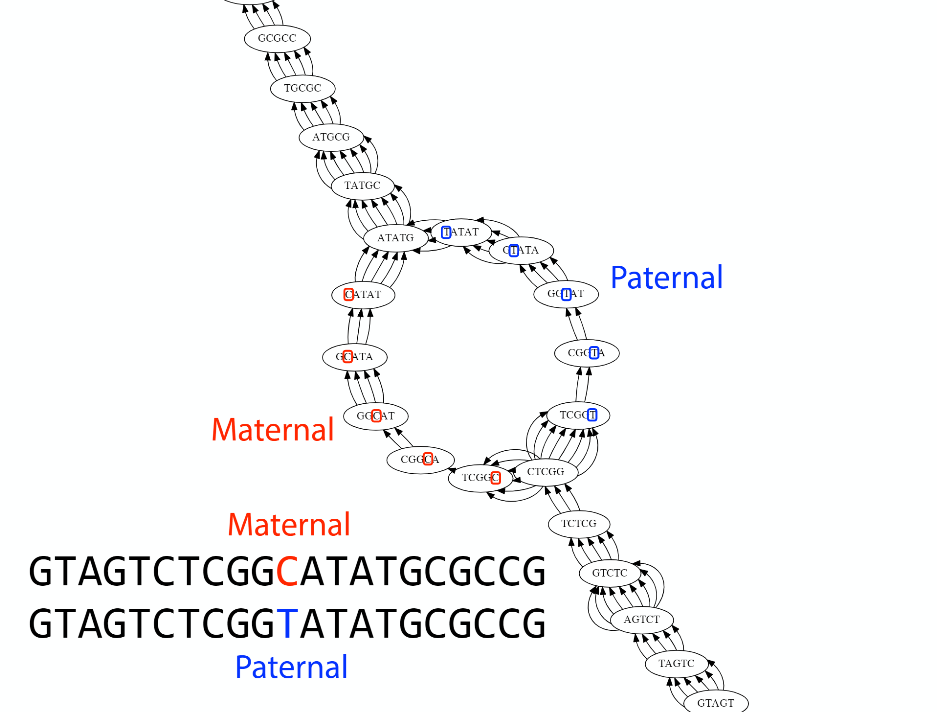
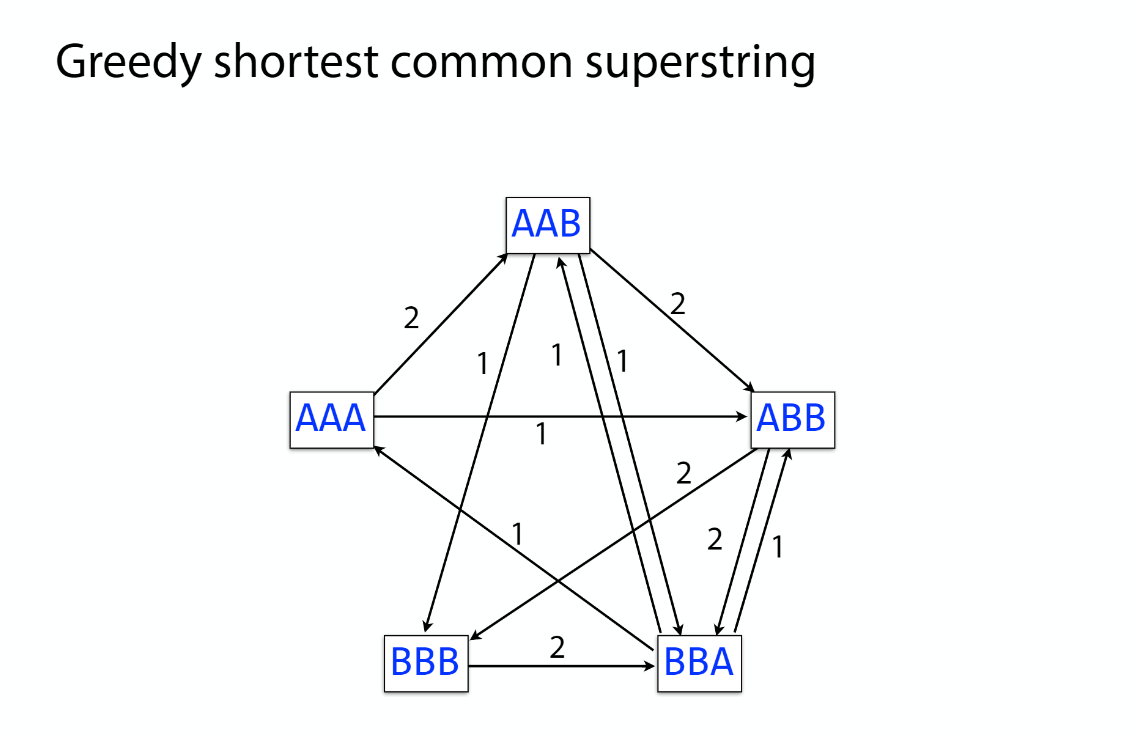
Transcriptome assembler
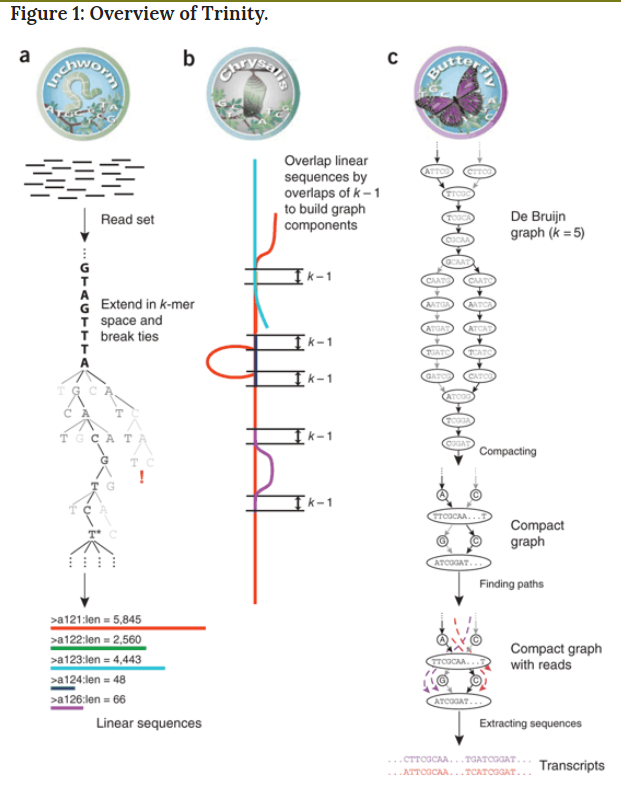
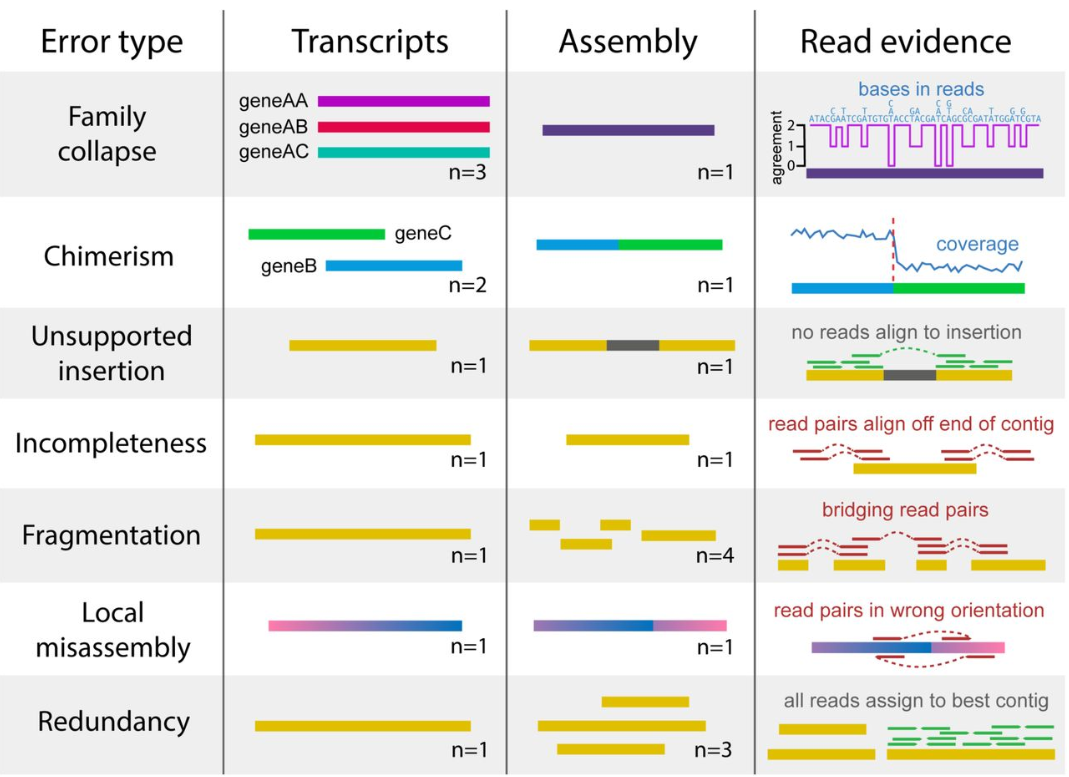
Transcriptome assembly error
Running Trinity
Trinity is run via the script: ‘Trinity’ found in the base installation directory.
Usage info is as follows:
conda create -n transcriptome_assembly
conda activate transcriptome_assembly
conda install -c bioconda/label/cf201901 trinity -y
conda install -c conda-forge/label/cf201901 nano -y
trinity
###############################################################################
#
# ______ ____ ____ ____ ____ ______ __ __
# | || \ | || \ | || || | |
# | || D ) | | | _ | | | | || | |
# |_| |_|| / | | | | | | | |_| |_|| ~ |
# | | | \ | | | | | | | | | |___, |
# | | | . \ | | | | | | | | | | |
# |__| |__|\_||____||__|__||____| |__| |____/
#
###############################################################################
#
# Required:
#
# --seqType <string> :type of reads: ( fa, or fq )
#
# --max_memory <string> :suggested max memory to use by Trinity where limiting can be enabled. (jellyfish, sorting, etc)
# provided in Gb of RAM, ie. '--max_memory 10G'
#
# If paired reads:
# --left <string> :left reads, one or more file names (separated by commas, not spaces)
# --right <string> :right reads, one or more file names (separated by commas, not spaces)
#
# Or, if unpaired reads:
# --single <string> :single reads, one or more file names, comma-delimited (note, if single file contains pairs, can use flag: --run_as_paired )
#
# Or,
# --samples_file <string> tab-delimited text file indicating biological replicate relationships.
# ex.
# cond_A cond_A_rep1 A_rep1_left.fq A_rep1_right.fq
# cond_A cond_A_rep2 A_rep2_left.fq A_rep2_right.fq
# cond_B cond_B_rep1 B_rep1_left.fq B_rep1_right.fq
# cond_B cond_B_rep2 B_rep2_left.fq B_rep2_right.fq
#
# # if single-end instead of paired-end, then leave the 4th column above empty.
#
####################################
## Misc: #########################
#
# --SS_lib_type <string> :Strand-specific RNA-Seq read orientation.
# if paired: RF or FR,
# if single: F or R. (dUTP method = RF)
# See web documentation.
#
# --CPU <int> :number of CPUs to use, default: 2
# --min_contig_length <int> :minimum assembled contig length to report
# (def=200)
#
# --long_reads <string> :fasta file containing error-corrected or circular consensus (CCS) pac bio reads
#
# --genome_guided_bam <string> :genome guided mode, provide path to coordinate-sorted bam file.
# (see genome-guided param section under --show_full_usage_info)
#
# --jaccard_clip :option, set if you have paired reads and
# you expect high gene density with UTR
# overlap (use FASTQ input file format
# for reads).
# (note: jaccard_clip is an expensive
# operation, so avoid using it unless
# necessary due to finding excessive fusion
# transcripts w/o it.)
#
# --trimmomatic :run Trimmomatic to quality trim reads
# see '--quality_trimming_params' under full usage info for tailored settings.
#
#
# --no_normalize_reads :Do *not* run in silico normalization of reads. Defaults to max. read coverage of 50.
# see '--normalize_max_read_cov' under full usage info for tailored settings.
# (note, as of Sept 21, 2016, normalization is on by default)
#
#
#
# --output <string> :name of directory for output (will be
# created if it doesn't already exist)
# default( your current working directory: "/Users/bhaas/GITHUB/trinityrnaseq/trinity_out_dir"
# note: must include 'trinity' in the name as a safety precaution! )
#
# --full_cleanup :only retain the Trinity fasta file, rename as ${output_dir}.Trinity.fasta
#
# --cite :show the Trinity literature citation
#
# --version :reports Trinity version (BLEEDING_EDGE) and exits.
#
# --show_full_usage_info :show the many many more options available for running Trinity (expert usage).
#
#
###############################################################################
#
# *Note, a typical Trinity command might be:
#
# Trinity --seqType fq --max_memory 50G --left reads_1.fq --right reads_2.fq --CPU 6
#
#
# and for Genome-guided Trinity:
#
# Trinity --genome_guided_bam rnaseq_alignments.csorted.bam --max_memory 50G
# --genome_guided_max_intron 10000 --CPU 6
#
# see: /Users/bhaas/GITHUB/trinityrnaseq/sample_data/test_Trinity_Assembly/
# for sample data and 'runMe.sh' for example Trinity execution
#
# For more details, visit: http://trinityrnaseq.github.io
#
###############################################################################
Trinity performs best with strand-specific data, in which case sense and antisense transcripts can be resolved. For protocols on strand-specific RNA-Seq, see: Borodina T, Adjaye J, Sultan M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol. 2011;500:79-98. PubMed PMID: 21943893.
If you have strand-specific data, specify the library type. There are four library types:
- Paired reads:
- RF: first read (/1) of fragment pair is sequenced as anti-sense (reverse(R)), and second read (/2) is in the sense strand (forward(F)); typical of the dUTP/UDG sequencing method.
- FR: first read (/1) of fragment pair is sequenced as sense (forward), and second read (/2) is in the antisense strand (reverse)
- Unpaired (single) reads:
- F: the single read is in the sense (forward) orientation
- R: the single read is in the antisense (reverse) orientation
By setting the –SS_lib_type parameter to one of the above, you are indicating that the reads are strand-specific. By default, reads are treated as not strand-specific.
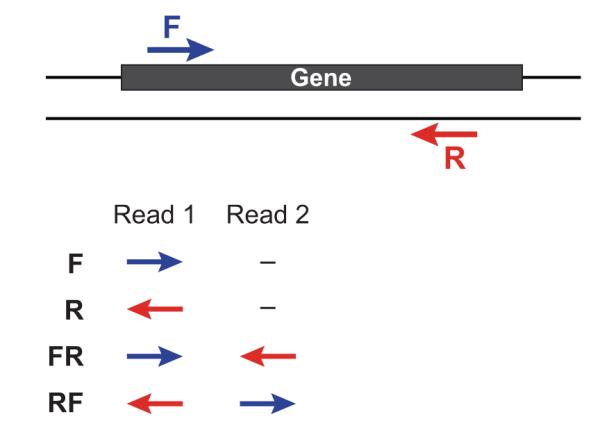
Other important considerations:
-
Whether you use Fastq or Fasta formatted input files, be sure to keep the reads oriented as they are reported by Illumina, if the data are strand-specific. This is because, Trinity will properly orient the sequences according to the specified library type. If the data are not strand-specific, no worries because the reads will be parsed in both orientations.
-
If you have both paired and unpaired data, and the data are NOT strand-specific, you can combine the unpaired data with the left reads of the paired fragments. Be sure that the unpaired reads have a /1 as a suffix to the accession value similarly to the left fragment reads. The right fragment reads should all have /2 as the accession suffix. Then, run Trinity using the –left and –right parameters as if all the data were paired.
-
If you have multiple paired-end library fragment sizes, set the ‘–group_pairs_distance’ according to the larger insert library. Pairings that exceed that distance will be treated as if they were unpaired by the Butterfly process.
-
by setting the ‘–CPU option’, you are indicating the maximum number of threads to be used by processes within Trinity. Note that Inchworm alone will be internally capped at 6 threads, since performance will not improve for this step beyond that setting)
Typical Trinity Command Line
A typical Trinity command for assembling non-strand-specific RNA-seq data would be like so, running the entire process on a single high-memory server (aim for ~1G RAM per ~1M ~76 base Illumina paired reads, but often much less memory is required):
Run Trinity like so:
Trinity --seqType fq --max_memory 50G \
--left reads_1.fq.gz --right reads_2.fq.gz --CPU 6
If you have multiple sets of fastq files, such as corresponding to multiple tissue types or conditions, etc., you can indicate them to Trinity like so:
Trinity --seqType fq --max_memory 50G \
--left condA_1.fq.gz,condB_1.fq.gz,condC_1.fq.gz \
--right condA_2.fq.gz,condB_2.fq.gz,condC_2.fq.gz \
--CPU 6
Paper need to read https://academic.oup.com/gigascience/article/8/5/giz039/5488105 https://www.nature.com/articles/nbt.1883 https://www.nature.com/articles/nprot.2013.084
Assignment
- Trimming Practice
- Make
Transcriptome_assemblyfolder under/data/gpfs/assoc/bch709-1/YOURID/BCH709_assignment - Change directory to the folder
Transcriptome_assembly - Copy all fastq.gz files in
/data/gpfs/assoc/bch709-1/Course_material/2020/RNASeq_raw_fastqtoTranscriptome_assemblyfolder
- Make
| Sample | Replication | Forward (read1) | Reverse (read2) |
|---|---|---|---|
| Wildtype | Rep1 | WT1_R1.fastq.gz | WT1_R2.fastq.gz |
| Wildtype | Rep2 | WT2_R1.fastq.gz | WT2_R2.fastq.gz |
| Wildtype | Rep3 | WT3_R1.fastq.gz | WT3_R2.fastq.gz |
| Stressed (drought) | Rep1 | DT1_R1.fastq.gz | DT1_R2.fastq.gz |
| Stressed (drought) | Rep2 | DT2_R1.fastq.gz | DT2_R2.fastq.gz |
| Stressed (drought) | Rep3 | DT3_R1.fastq.gz | DT3_R2.fastq.gz |
- Run
Trim-Galoreand processMultiQC - Generate 6 trimmed output from
MultiQCand uploadhtmlfile to WebCanvas.
** IF YOU USE “LOOP” FOR THIS JOB, PLEASE ATTACH YOUR COMMAND, YOU WILL GET ADDITIONAL 10 POINTS **
Until 10/19/20 9:00AM Late submission will not be allowed.
Reference:
- Conda documentation https://docs.conda.io/en/latest/
- Conda-forge https://conda-forge.github.io/
- BioConda https://bioconda.github.io/
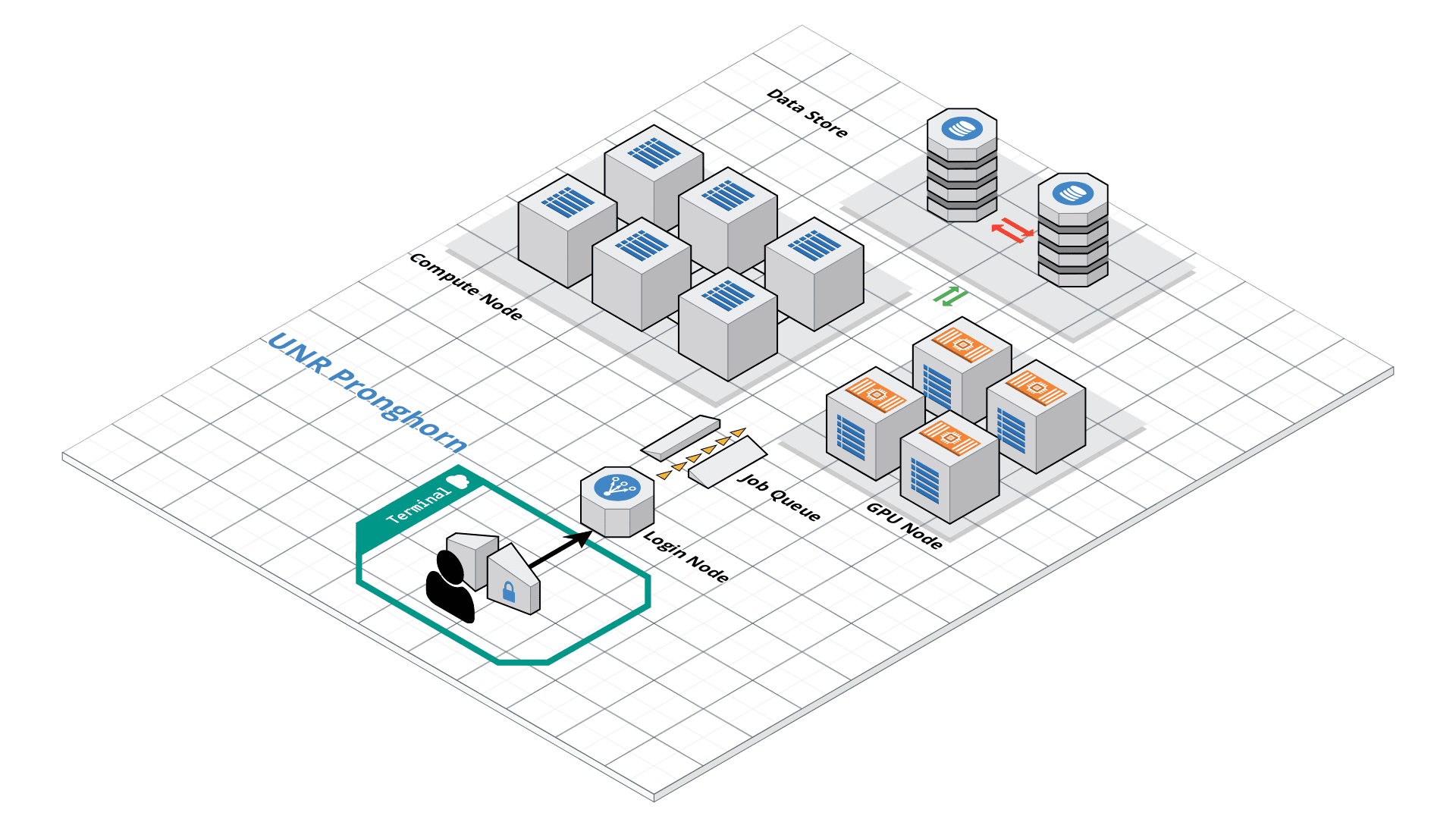
Slurm Quick Start Tutorial
Resource sharing on a supercomputer dedicated to technical and/or scientific computing is often organized by a piece of software called a resource manager or job scheduler. Users submit jobs, which are scheduled and allocated resources (CPU time, memory, etc.) by the resource manager.
Slurm is a resource manager and job scheduler designed to do just that, and much more. It was originally created by people at the Livermore Computing Center, and has grown into a full-fledge open-source software backed up by a large community, commercially supported by the original developers, and installed in many of the Top500 supercomputers.
Gathering information Slurm offers many commands you can use to interact with the system. For instance, the sinfo command gives an overview of the resources offered by the cluster, while the squeue command shows to which jobs those resources are currently allocated.
By default, sinfo lists the partitions that are available. A partition is a set of compute nodes (computers dedicated to… computing) grouped logically. Typical examples include partitions dedicated to batch processing, debugging, post processing, or visualization.
sinfo
Show the State of Nodes
sinfo --all
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
cpu-s2-core-0 up 14-00:00:0 2 mix cpu-[8-9]
cpu-s2-core-0 up 14-00:00:0 7 alloc cpu-[1-2,4-6,78-79]
cpu-s2-core-0 up 14-00:00:0 44 idle cpu-[0,3,7,10-47,64,76-77]
cpu-s3-core-0* up 2:00:00 2 mix cpu-[8-9]
cpu-s3-core-0* up 2:00:00 7 alloc cpu-[1-2,4-6,78-79]
cpu-s3-core-0* up 2:00:00 44 idle cpu-[0,3,7,10-47,64,76-77]
gpu-s2-core-0 up 14-00:00:0 11 idle gpu-[0-10]
cpu-s6-test-0 up 15:00 2 idle cpu-[65-66]
cpu-s1-pgl-0 up 14-00:00:0 1 mix cpu-49
cpu-s1-pgl-0 up 14-00:00:0 1 alloc cpu-48
cpu-s1-pgl-0 up 14-00:00:0 2 idle cpu-[50-51]
In the above example, we see two partitions, named batch and debug. The latter is the default partition as it is marked with an asterisk. All nodes of the debug partition are idle, while two of the batch partition are being used.
The sinfo command also lists the time limit (column TIMELIMIT) to which jobs are subject. On every cluster, jobs are limited to a maximum run time, to allow job rotation and let every user a chance to see their job being started. Generally, the larger the cluster, the smaller the maximum allowed time. You can find the details on the cluster page.
You can actually specify precisely what information you would like sinfo to output by using its –format argument. For more details, have a look at the command manpage with man sinfo.
sinfo advance
sinfo $OPTIONS -o "%13n %8t %4c %8z %15C %8O %8m %8G %18P %f" | egrep s2
squeue
The squeue command shows the list of jobs which are currently running (they are in the RUNNING state, noted as ‘R’) or waiting for resources (noted as ‘PD’, short for PENDING).
squeue
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
983204 cpu-s2-co neb_K jzhang23 R 6-09:05:47 1 cpu-6
983660 cpu-s2-co RT3.sl yinghanc R 12:56:17 1 cpu-9
983659 cpu-s2-co RT4.sl yinghanc R 12:56:21 1 cpu-8
983068 cpu-s2-co Gd-bound dcantu R 7-06:16:01 2 cpu-[78-79]
983067 cpu-s2-co Gd-unbou dcantu R 1-17:41:56 2 cpu-[1-2]
983472 cpu-s2-co ub-all dcantu R 3-10:05:01 2 cpu-[4-5]
982604 cpu-s1-pg wrap wyim R 12-14:35:23 1 cpu-49
983585 cpu-s1-pg wrap wyim R 1-06:28:29 1 cpu-48
983628 cpu-s1-pg wrap wyim R 13:44:46 1 cpu-49
SBATCH
Now the question is: How do you create a job?
A job consists in two parts: resource requests and job steps. Resource requests consist in a number of CPUs, computing expected duration, amounts of RAM or disk space, etc. Job steps describe tasks that must be done, software which must be run.
The typical way of creating a job is to write a submission script. A submission script is a shell script, e.g. a Bash script, whose comments, if they are prefixed with SBATCH, are understood by Slurm as parameters describing resource requests and other submissions options. You can get the complete list of parameters from the sbatch manpage man sbatch.
Important
The SBATCH directives must appear at the top of the submission file, before any other line except for the very first line which should be the shebang (e.g. #!/bin/bash). The script itself is a job step. Other job steps are created with the srun command. For instance, the following script, hypothetically named submit.sh,
nano setting
set nowrap
set softwrap
set const
## Nanorc files
include "/usr/share/nano/nanorc.nanorc"
## C/C++
include "/usr/share/nano/c.nanorc"
## HTML
include "/usr/share/nano/html.nanorc"
## TeX
include "/usr/share/nano/tex.nanorc"
## Quoted emails (under e.g. mutt)
include "/usr/share/nano/mutt.nanorc"
## Patch files
include "/usr/share/nano/patch.nanorc"
## Manpages
include "/usr/share/nano/man.nanorc"
## Groff
include "/usr/share/nano/groff.nanorc"
## Perl
include "/usr/share/nano/perl.nanorc"
## Python
include "/usr/share/nano/python.nanorc"
## Ruby
include "/usr/share/nano/ruby.nanorc"
## Java
include "/usr/share/nano/java.nanorc"
## Assembler
include "/usr/share/nano/asm.nanorc"
## Bourne shell scripts
include "/usr/share/nano/sh.nanorc"
## POV-Ray
include "/usr/share/nano/pov.nanorc"
nano submit.sh
#!/bin/bash
#SBATCH --job-name=test
#SBATCH --cpus-per-task=16
#SBATCH --time=10:00
#SBATCH --account=cpu-s2-bch709-1
#SBATCH --partition=cpu-s2-core-0
#SBATCH --mem-per-cpu=1g
#SBATCH --mail-type=begin
#SBATCH --mail-type=end
#SBATCH --mail-type=fail
#SBATCH --mail-user=<YOUR ID>@unr.edu
echo "Hello Pronghorn"
seq 1 8000
would request one CPU for 10 minutes, along with 1g of RAM, in the default queue. When started, the job would run a first job step srun hostname, which will launch the UNIX command hostname on the node on which the requested CPU was allocated. Then, a second job step will start the sleep command. Note that the –job-name parameter allows giving a meaningful name to the job and the –output parameter defines the file to which the output of the job must be sent.
Once the submission script is written properly, you need to submit it to slurm through the sbatch command, which, upon success, responds with the jobid attributed to the job. (The dollar sign below is the shell prompt)
$ chmod 775 submit.sh
$ sbatch submit.sh
sbatch: Submitted batch job ########
Conda env
conda activate rnaseq
File prepare
mkdir -p /data/gpfs/assoc/bch709-1/<YOURID>/rnaseq_slurm/fastq
cd /data/gpfs/assoc/bch709-1/<YOURID>/rnaseq_slurm/fastq
cp /data/gpfs/assoc/bch709-1/Course_material/2020/RNASeq_raw_fastq/*.gz .
cd ../
Run trimming
pwd
nano trim.sh
#!/bin/bash
#SBATCH --job-name=<TRIM>
#SBATCH --time=4:00:00
#SBATCH --account=cpu-s2-bch709-0
#SBATCH --partition=cpu-s2-core-0
#SBATCH --cpus-per-task=8
#SBATCH --mem-per-cpu=32g
#SBATCH --mail-type=fail
#SBATCH --mail-type=begin
#SBATCH --mail-type=end
#SBATCH --mail-user=<YOUR ID>@unr.edu
#SBATCH --output=<TRIM>.out
trim_galore --paired --three_prime_clip_R1 10 --three_prime_clip_R2 10 --cores 8 --max_n 40 --gzip -o trim /data/gpfs/assoc/bch709-1/<YOURID>/rnaseq_slurm/fastq/DT1_R1.fastq.gz /data/gpfs/assoc/bch709-1/<YOURID>/rnaseq_slurm/fastq/DT1_R2.fastq.gz
trim_galore --paired --three_prime_clip_R1 10 --three_prime_clip_R2 10 --cores 8 --max_n 40 --gzip -o trim /data/gpfs/assoc/bch709-1/<YOURID>/rnaseq_slurm/fastq/DT2_R1.fastq.gz /data/gpfs/assoc/bch709-1/<YOURID>/rnaseq_slurm/fastq/DT2_R2.fastq.gz
.
.
.
.
.
.
Submit job
chmod 775 trim.sh
sbatch trim.sh
PLEASE CHECK YOUR MULTIQC after job running
MultiQC
multiqc --filename trim .
Trinity run
Need to activate Transcriptome assembly environment
nano trinity.sh
#!/bin/bash
#SBATCH --job-name=<TRINITY>
#SBATCH --time=6:00:00
#SBATCH --account=cpu-s2-bch709-1
#SBATCH --partition=cpu-s2-core-0
#SBATCH --cpus-per-task=24
#SBATCH --mem=60g
#SBATCH --mail-type=fail
#SBATCH --mail-type=begin
#SBATCH --mail-type=end
#SBATCH --mail-user=<YOUR ID>@unr.edu
#SBATCH --output=<TRINITY>.out
Trinity --seqType fq --CPU 4 --max_memory 60G --left trim/DT1_R1_val_1.fq.gz,trim/DT2_R1_val_1.fq.gz,trim/DT3_R1_val_1.fq.gz,trim/WT1_R1_val_1.fq.gz,trim/WT2_R1_val_1.fq.gz,trim/WT3_R1_val_1.fq.gz --right trim/DT1_R2_val_2.fq.gz,trim/DT2_R2_val_2.fq.gz,trim/DT3_R2_val_2.fq.gz,trim/WT1_R2_val_2.fq.gz,trim/WT2_R2_val_2.fq.gz,trim/WT3_R2_val_2.fq.gz
Please consider relative path and absolute path
Submit job
chmod 775 trinity.sh
sbatch trinity.sh
MultiQC
multiqc . -n assembly
Use SCP for downloading and send me the file
