Pairwise alignment
The process or result of matching up the nucleotide or amino acid residues of two or more biological sequences to achieve maximal levels of identity and, in the case of amino acid sequences, conservation, for the purpose of assessing the degree of similarity and the possibility of homology.
One of the most fundamental problems in bioinformatics is determining how “similar” a pair of biological sequences are. There are many applications for this, including inferring the function or source organism of an unknown gene sequence, developing hypotheses about the relatedness of organisms, or grouping sequences from closely related organisms. On the surface this seems like a pretty straight-forward problem, not one that would have been at the center of decades of research and the subject of one of the most cited papers in modern biology. In this chapter we’ll explore why determining sequence similarity is harder than it might initially seem, and learn about pairwise sequence alignment, the standard approach for determining sequence similarity.
Protein vs. DNA sequence alignment
Protein amino acid sequences are preferred over DNA sequences for a list of reasons.
- Protein residues are more informative - a change in DNA (especially the 3rd position) does not necessarily change the AA.
- The larger number of amino acids than nucleic acids makes it easier to find significance.
- Some amino acids share related biochemical properties, which can be accounted for when scoring multiple pairwise alignments.
- Protein sequence comparisons can link back to over a billion years ago, whereas DNA sequence comparisons can only go back up to 600 mya. Thus, protein sequences are far better for evolutionary studies.
However, there are some obvious instances when DNA alignments are needed.
- When confirming the identity of cDNA (forensic sequencing).
- When studying noncoding regions of DNA. These regions evolve at a faster rate than coding DNA, while mitochondrial noncoding DNA evolves even faster.
- When studying DNA mutations.
- When researching on very similar organisms such as Neanderthals and modern humans.
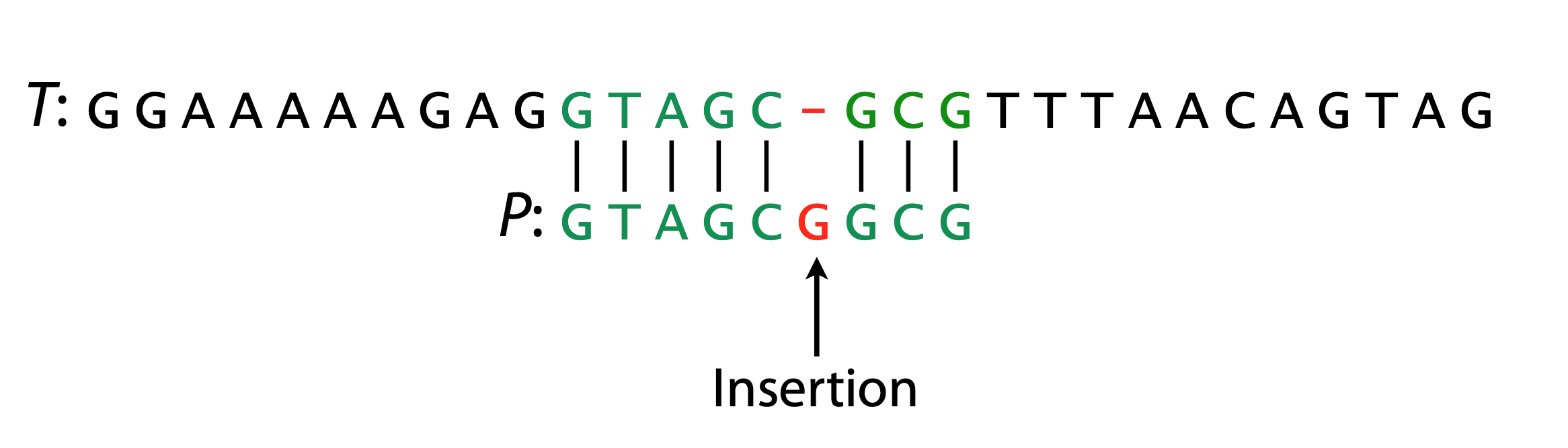
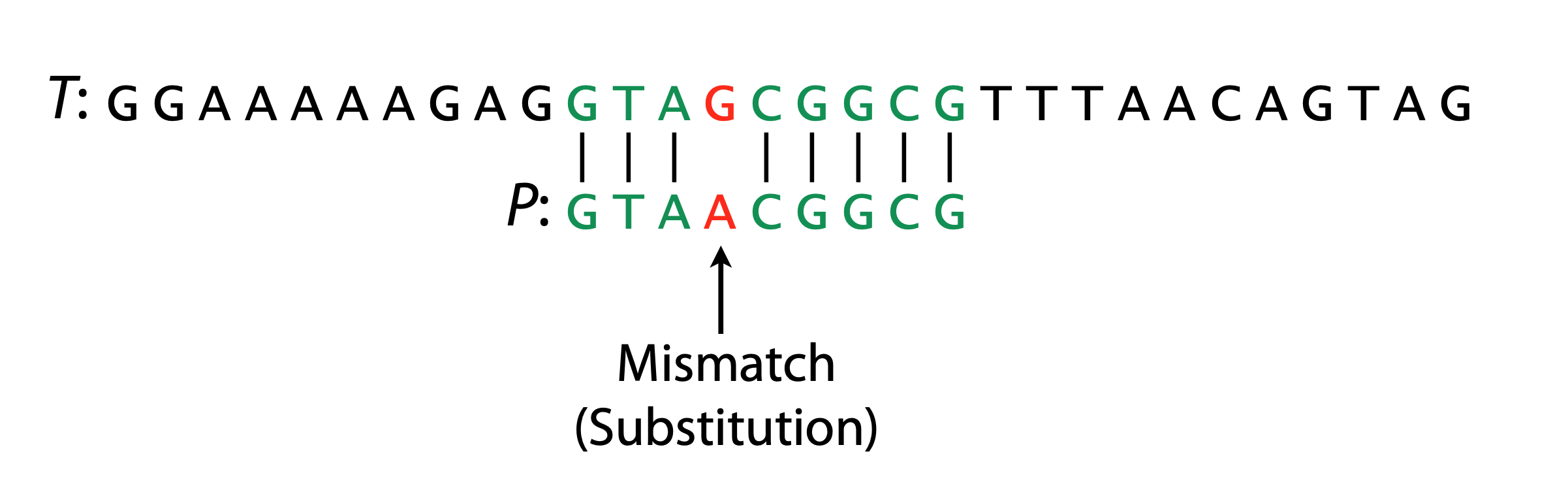
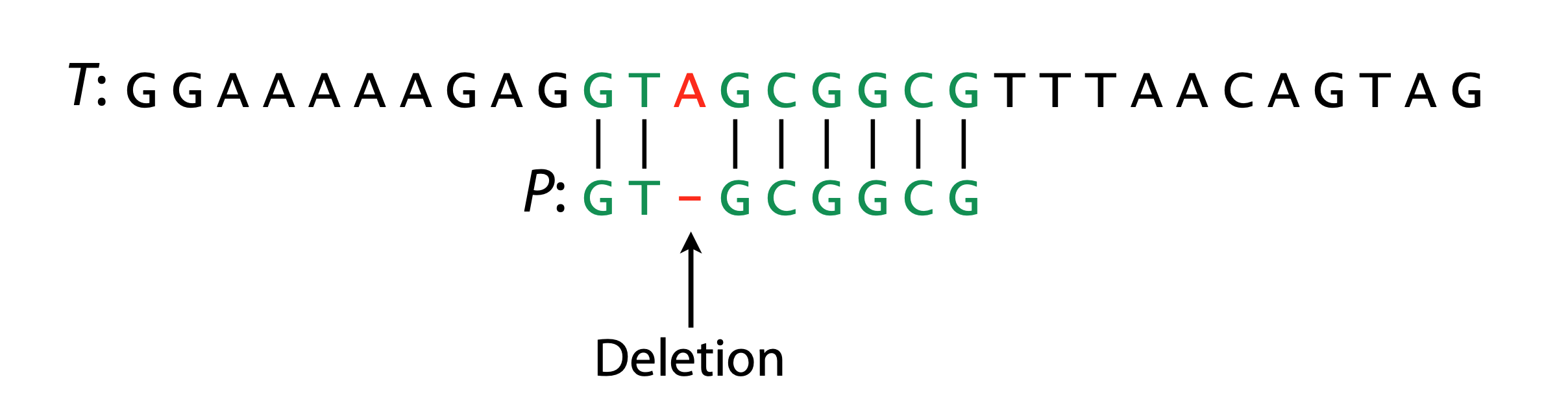
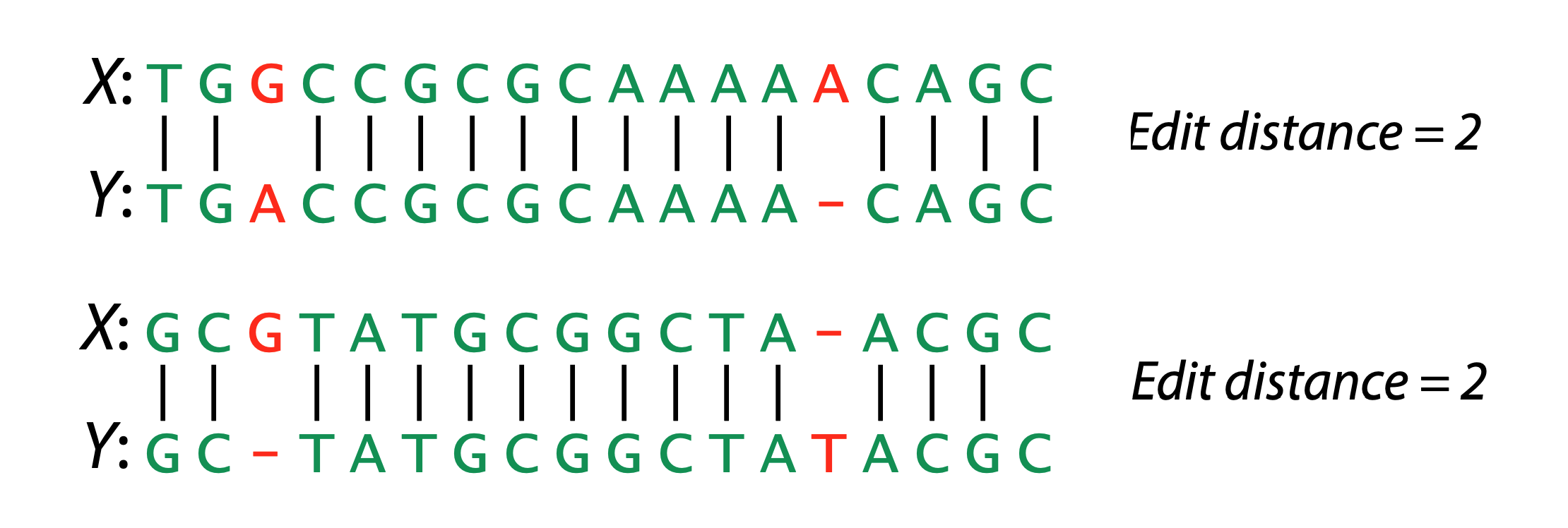
Hamming Distance
Metric to compare sequences (DNA, AA, ASCII, binary, etc…)
- Non-negative, identity, symmetry, triangle equality
- How many characters are different between the 2 strings?
- Minimum number of substitutions required to change transform A into B
- Traditionally defined for end-to-end comparisons

Here end-to-end (global) for query, partial (local) for reference
- Find all occurrences of GATTACA with Hamming Distance ≤ 1
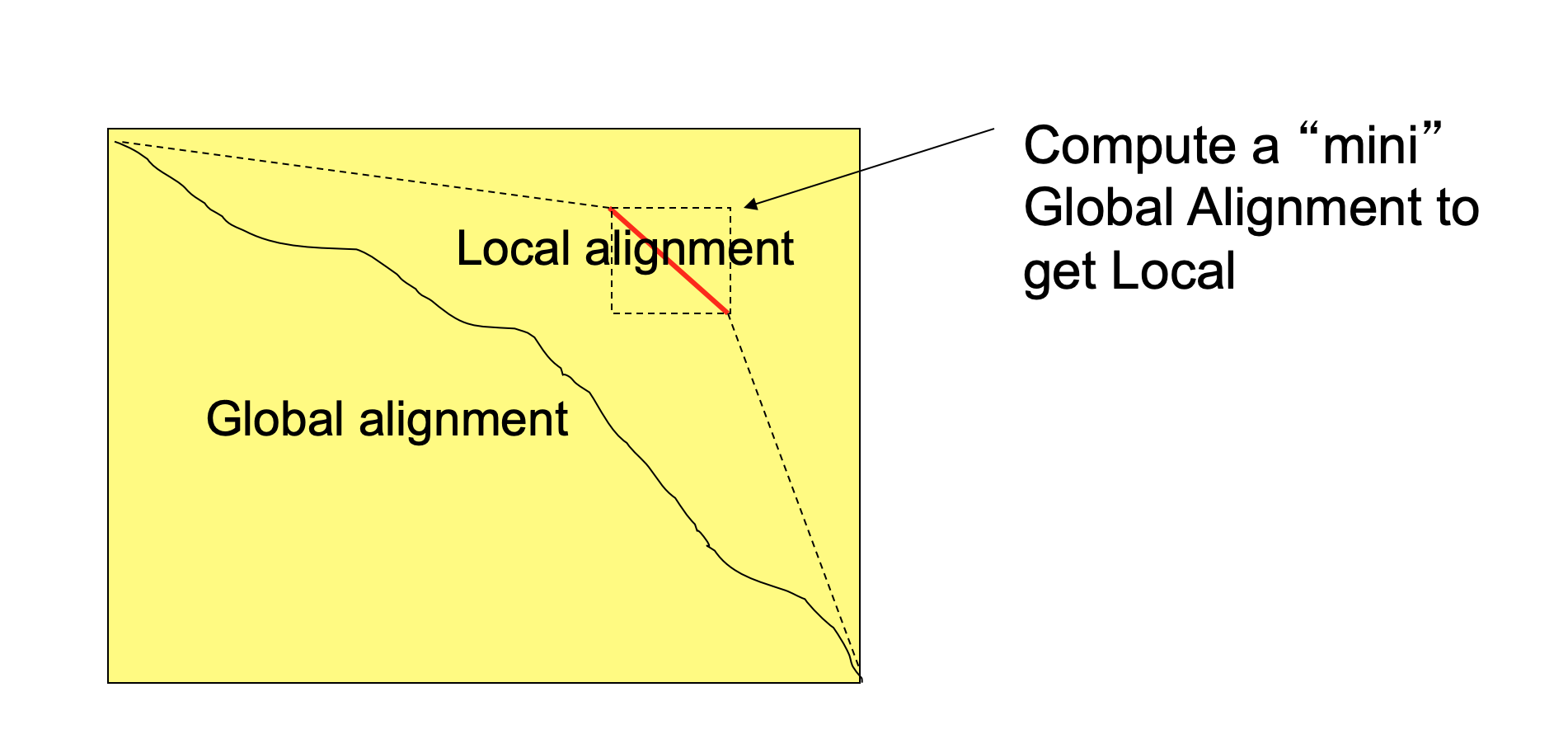
Local vs Global alignment
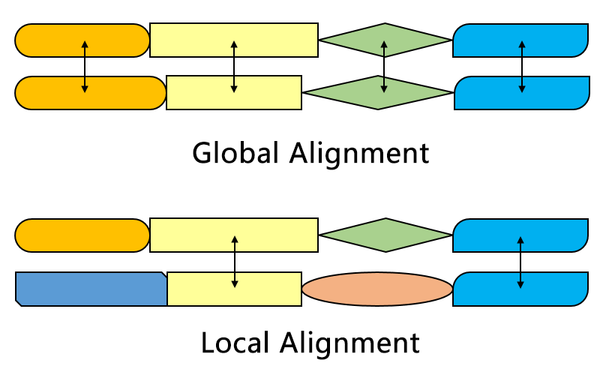
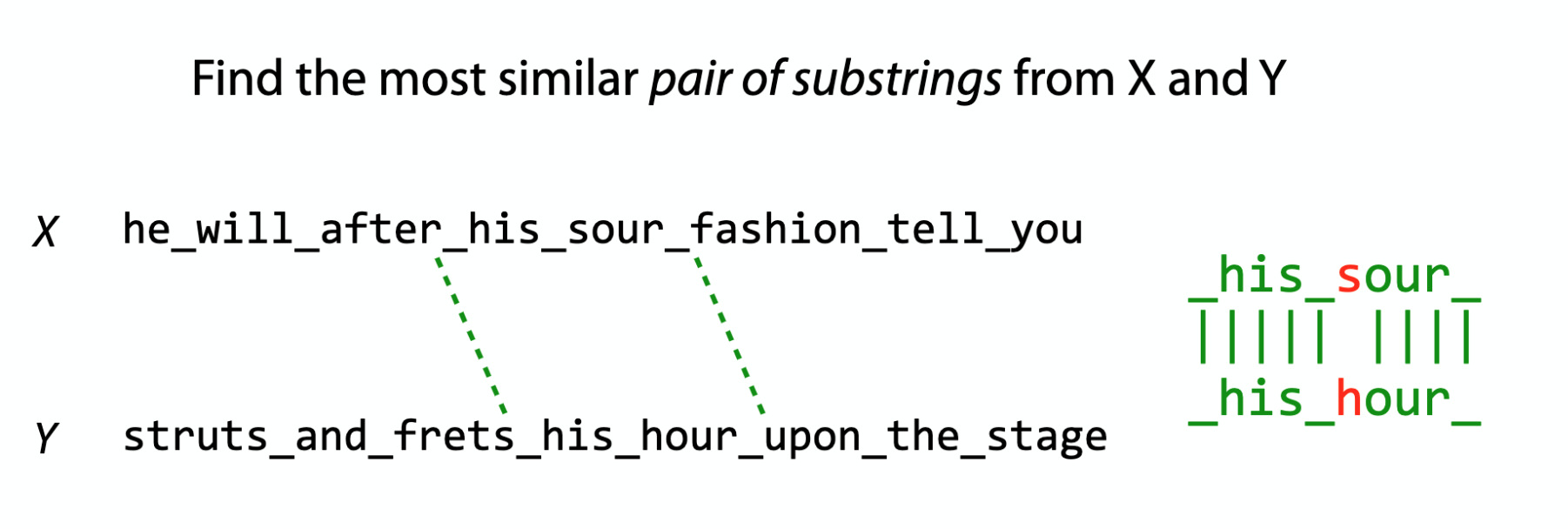
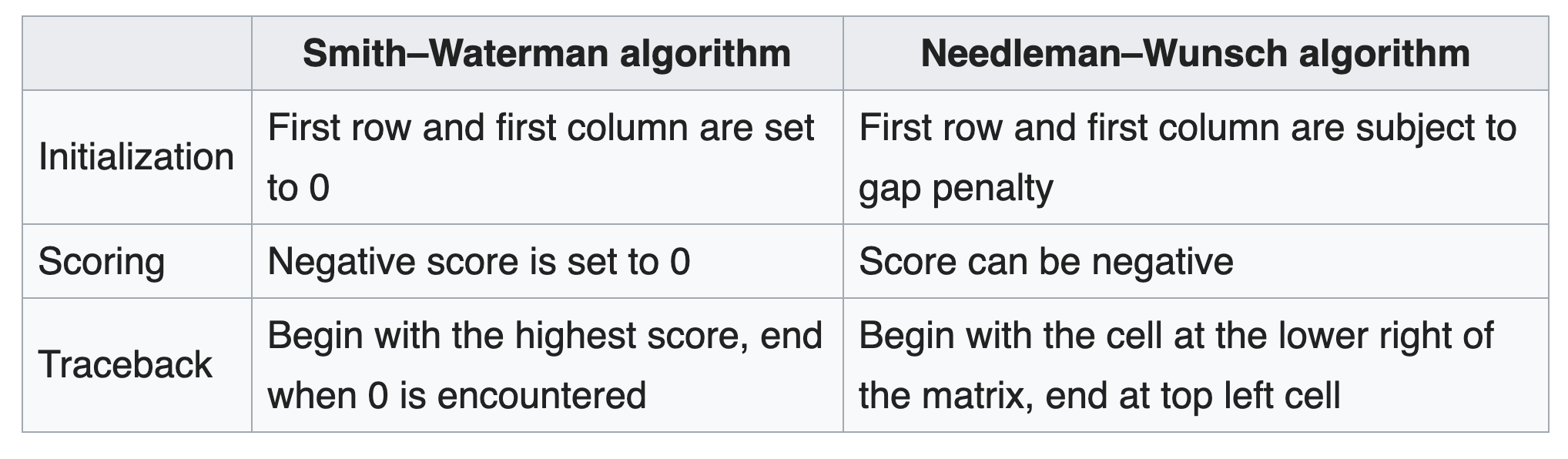
The Smith–Waterman algorithm finds the segments in two sequences that have similarities while the Needleman–Wunsch algorithm aligns two complete sequences. Therefore, they serve different purposes. Both algorithms use the concepts of a substitution matrix, a gap penalty function, a scoring matrix, and a traceback process.

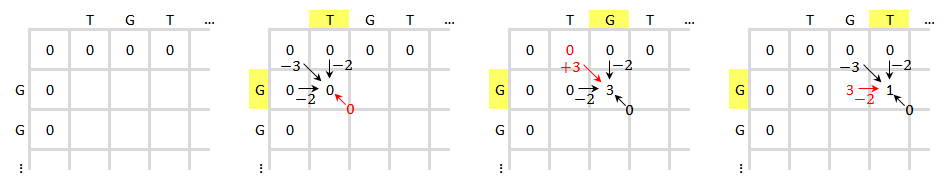
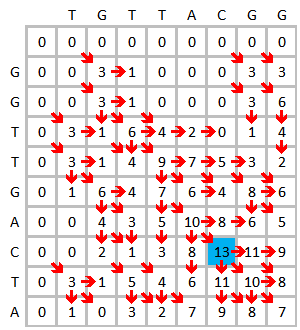
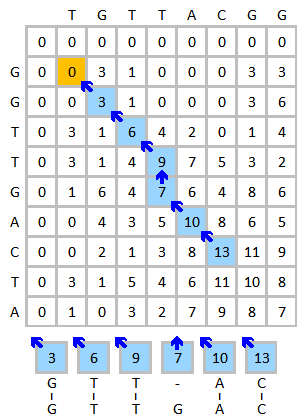
Take the alignment of DNA sequences TGTTACGG and GGTTGACTA as an example. Use the following scheme:
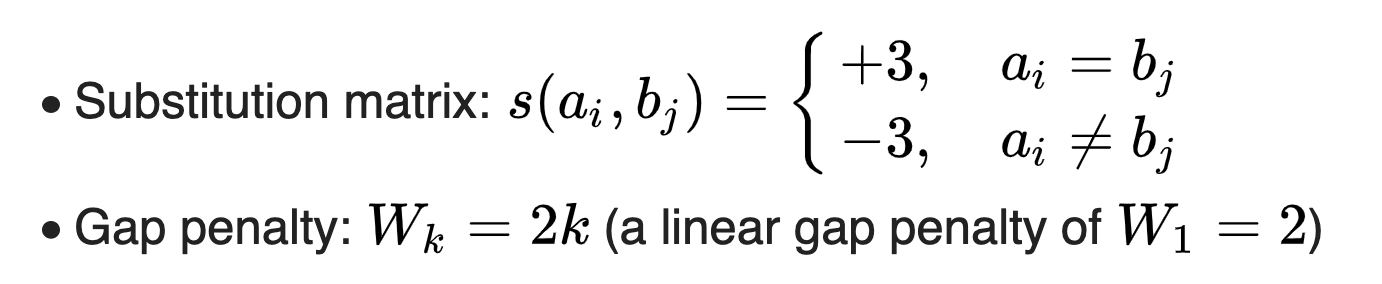
Initialize and fill the scoring matrix, shown as below. This figure shows the scoring process of the first three elements. The yellow color indicates the bases that are being considered. The red color indicates the highest possible score for the cell being scored.
location
mkdir /data/gpfs/assoc/bch709/<YOURID>/alignment
cd $!
Install software
create alignment env, activate and install blast muscle clustalo from bioconda
Actual Alignment
Match score: 3
Mismatch score: -3
Gap penalty: -2
TTAGTAT
TAGCTAT
| T | T | A | G | T | A | T | |
|---|---|---|---|---|---|---|---|
| T | |||||||
| A | |||||||
| G | |||||||
| C | |||||||
| T | |||||||
| A | |||||||
| T |
Example Protein sequences
- save it as example.aa
>sp|P10242|MYB_HUMAN Transcriptional activator Myb OS=Homo sapiens OX=9606 GN=MYB PE=1 SV=2
MARRPRHSIYSSDEDDEDFEMCDHDYDGLLPKSGKRHLGKTRWTREEDEKLKKLVEQNGT
DDWKVIANYLPNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIELVQKYGPKRWSVIAK
HLKGRIGKQCRERWHNHLNPEVKKTSWTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNA
IKNHWNSTMRRKVEQEGYLQESSKASQPAVATSFQKNSHLMGFAQAPPTAQLPATGQPTV
NNDYSYYHISEAQNVSSHVPYPVALHVNIVNVPQPAAAAIQRHYNDEDPEKEKRIKELEL
LLMSTENELKGQQVLPTQNHTCSYPGWHSTTIADHTRPHGDSAPVSCLGEHHSTPSLPAD
PGSLPEESASPARCMIVHQGTILDNVKNLLEFAETLQFIDSFLNTSSNHENSDLEMPSLT
STPLIGHKLTVTTPFHRDQTVKTQKENTVFRTPAIKRSILESSPRTPTPFKHALAAQEIK
YGPLKMLPQTPSHLVEDLQDVIKQESDESGIVAEFQENGPPLLKKIKQEVESPTDKSGNF
FCSHHWEGDSLNTQLFTQTSPVADAPNILTSSVLMAPASEDEDNVLKAFTVPKNRSLASP
LQPCSSTWEPASCGKMEEQMTSSSQARKYVNAFSARTLVM
>sp|P46200|MYB_BOVIN Transcriptional activator Myb OS=Bos taurus OX=9913 GN=MYB PE=2 SV=1
MARRPRHSIYSSDEDDEDIEMCDHDYDGLLPKSGKRHLGKTRWTREEDEKLKKLVEQNGT
DDWKVIANYLPNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIELVQKYGPKRWSVIAK
HLKGRIGKQCRERWHNHLNPEVKKTSWTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNA
IKNHWNSTMRRKVEQEGYLQESSKASQPAVTTSFQKNSHLMGFTHAPPSAQLPPAGQPSV
NSDYPYYHISEAQNVSSHVPYPVALHVNIVNVPQPAAAAIQRHYNDEDPEKEKRIKELEL
LLMSTENELKGQQALPTQNHTCSYPGWHSTTIADHTRPHGDSAPVSCLEEHHSTPSLPAD
PGSLPEESASPARCMIFHQSTILDNVKNLLEFAETLQFIDSFLNTSNNHENLDLEMPSLT
STPLNGHKLTVTTPFHRDQTVKIQKENTIFRTPAIKRSILEGSPRTPTPFKHALTAQEIK
YGPLKMLPQTPSHLVEDLQDEIKQESDESGIVAEFQENGQPLLKKIKQEVESPTDKAGNF
FCSNHWEGDSLNTQLFTQASPVADMPNILTSSVLMTPVSEDEDNVLKAFTVPKSRSLASP
LQPCNGAWESASCGKTDDQMTASGQSRKYVNAFSTRTLVM
>tr|A0A2I2ZIL5|A0A2I2ZIL5_GORGO MYB proto-oncogene, transcription factor OS=Gorilla gorilla gorilla OX=9595 PE=4 SV=1
MARRPRHSIYSSDEDDEDFEMCDHDYDGLLPKSGKRHLGKTRWTREEDEKLKKLVEQNGT
DDWKVIANYLPNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIELVQKYGPKRWSVIAK
HLKGRIGKQCRERWHNHLNPEVKKTSWTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNA
IKNHWNSTMRRKVEQEGYLQESSKASQPAVATSFQKNSHLMGFAQAPPTAQLPATGQPTV
NNDYSYYHISEAQNVSSHVPYPVALHVNIVNVPQPAAAAIQRHYNDEDPEKEKRIKELEL
LLMSTENELKGQQVLPTQNHTCSYPGWHSTTIADHTRPHGDSAPVSCLGEHHSTPSLPAD
PGSLPEESASPARCMIVHQGTILDNVKNLLEFAETLQFIDSFLNTSSNHENSDLEMPSLT
STPLIGHKLTVTTPFHRDQTVKTQKENTVFRTPAIKRSILESSPRTPTPFKHALAAQEIK
YGPLKMLPQTPSHLVEDLQDVIKQESDESGIVAEFQENGPPLLKKIKQEVESPTDKSGNF
FCSHHWEGDGLNTQLFTQTSPVADAPNILTSSVLMAPASEDEDNVLKAFTVPKNRSLASP
LQPCSSTWEPASCGKLEEQMTASSQARKYVNAFSARTLVM
>XP_012418097.1 PREDICTED: transcriptional activator Myb isoform X5 [Odobenus rosmarus divergens]
MARRPRHSIYSSDEDDEDIEMCDHDYDGLLPKSGKRHLGKTRWTREEDEKLKKLVEQNGTDDWKVIANYL
PNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHLKGRIGKQCRERWHNHLNP
EVKKTSWTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEGYLQESSKASPPTV
ATSFQKNSHLMGFAHAPPSAHLPPAGQPSVNNDYSYYHISEAQNVSSHVPYPVALHVNIVNVPQPAAAAI
QRHYNDEDPEKEKRIKELELLLMSTENELKGQQTQNHTCSYPGWHSTTIADHTRPHGDSAPVSCLEEHHS
TPSLPVDPGSLPEESASPARCMIVHQGTILDNVKNLLEFAETLQFIDSFLNTSGNHENLDLEMPSLTSTP
LNGHKLTVTTPFHRDQTVKTQKENTIFRTPAIKRSILESSPRTPTPFKHGLAAQEIKYGPLKMLPQTPSH
LVEDLQDVIKQESDEPGIVAEFQENGPPLLKKIKQEVESPTDKAGNFFCSSHWEGESLNTQLFPQALPVT
DVPNILTSSVLMTPVSEDEDNVLKAFTVPKNRSLASPLQPCGGAWEAASCGKTEDQMTASGQARKYVNAF
STRTLVM
Example Nucleotide Sequence
- save it as example.fasta
>NM_001130173.1
ATGGCCCGAAGACCCCGGCACAGCATATATAGCAGTGACGAGGATGATGAGGACTTTGAGATGTGTGACC
ATGACTATGATGGGCTGCTTCCCAAGTCTGGAAAGCGTCACTTGGGGAAAACAAGGTGGACCCGGGAAGA
GGATGAAAAACTGAAGAAGCTGGTGGAACAGAATGGAACAGATGACTGGAAAGTTATTGCCAATTATCTC
CCGAATCGAACAGATGTGCAGTGCCAGCACCGATGGCAGAAAGTACTAAACCCTGAGCTCATCAAGGGTC
CTTGGACCAAAGAAGAAGATCAGAGAGTGATAGAGCTTGTACAGAAATACGGTCCGAAACGTTGGTCTGT
TATTGCCAAGCACTTAAAGGGGAGAATTGGAAAACAATGTAGGGAGAGGTGGCATAACCACTTGAATCCA
GAAGTTAAGAAAACCTCCTGGACAGAAGAGGAAGACAGAATTATTTACCAGGCACACAAGAGACTGGGGA
ACAGATGGGCAGAAATCGCAAAGCTACTGCCTGGACGAACTGATAATGCTATCAAGAACCACTGGAATTC
TACAATGCGTCGGAAGGTCGAACAGGAAGGTTATCTGCAGGAGTCTTCAAAAGCCAGCCAGCCAGCAGTG
GCCACAAGCTTCCAGAAGAACAGTCATTTGATGGGTTTTGCTCAGGCTCCGCCTACAGCTCAACTCCCTG
CCACTGGCCAGCCCACTGTTAACAACGACTATTCCTATTACCACATTTCTGAAGCACAAAATGTCTCCAG
TCATGTTCCATACCCTGTAGCGTTACATGTAAATATAGTCAATGTCCCTCAGCCAGCTGCCGCAGCCATT
CAGAGACACTATAATGATGAAGACCCTGAGAAGGAAAAGCGAATAAAGGAATTAGAATTGCTCCTAATGT
CAACCGAGAATGAGCTAAAAGGACAGCAGACACAGAACCACACATGCAGCTACCCCGGGTGGCACAGCAC
CACCATTGCCGACCACACCAGACCTCATGGAGACAGTGCACCTGTTTCCTGTTTGGGAGAACACCACTCC
ACTCCATCTCTGCCAGCGGATCCTGGCTCCCTACCTGAAGAAAGCGCCTCGCCAGCAAGGTGCATGATCG
TCCACCAGGGCACCATTCTGGATAATGTTAAGAACCTCTTAGAATTTGCAGAAACACTCCAATTTATAGA
TTCTTTCTTAAACACTTCCAGTAACCATGAAAACTCAGACTTGGAAATGCCTTCTTTAACTTCCACCCCC
CTCATTGGTCACAAATTGACTGTTACAACACCATTTCATAGAGACCAGACTGTGAAAACTCAAAAGGAAA
ATACTGTTTTTAGAACCCCAGCTATCAAAAGGTCAATCTTAGAAAGCTCTCCAAGAACTCCTACACCATT
CAAACATGCACTTGCAGCTCAAGAAATTAAATACGGTCCCCTGAAGATGCTACCTCAGACACCCTCTCAT
CTAGTAGAAGATCTGCAGGATGTGATCAAACAGGAATCTGATGAATCTGGAATTGTTGCTGAGTTTCAAG
AAAATGGACCACCCTTACTGAAGAAAATCAAACAAGAGGTGGAATCTCCAACTGATAAATCAGGAAACTT
CTTCTGCTCACACCACTGGGAAGGGGACAGTCTGAATACCCAACTGTTCACGCAGACCTCGCCTGTGGCA
GATGCACCGAATATTCTTACAAGCTCCGTTTTAATGGCACCAGCATCAGAAGATGAAGACAATGTTCTCA
AAGCATTTACAGTACCTAAAAACAGGTCCCTGGCGAGCCCCTTGCAGCCTTGTAGCAGTACCTGGGAACC
TGCATCCTGTGGAAAGATGGAGGAGCAGATGACATCTTCCAGTCAAGCTCGTAAATACGTGAATGCATTC
TCAGCCCGGACGCTGGTCATGTGA
>NM_005375.3
ATGGCCCGAAGACCCCGGCACAGCATATATAGCAGTGACGAGGATGATGAGGACTTTGAGATGTGTGACC
ATGACTATGATGGGCTGCTTCCCAAGTCTGGAAAGCGTCACTTGGGGAAAACAAGGTGGACCCGGGAAGA
GGATGAAAAACTGAAGAAGCTGGTGGAACAGAATGGAACAGATGACTGGAAAGTTATTGCCAATTATCTC
CCGAATCGAACAGATGTGCAGTGCCAGCACCGATGGCAGAAAGTACTAAACCCTGAGCTCATCAAGGGTC
CTTGGACCAAAGAAGAAGATCAGAGAGTGATAGAGCTTGTACAGAAATACGGTCCGAAACGTTGGTCTGT
TATTGCCAAGCACTTAAAGGGGAGAATTGGAAAACAATGTAGGGAGAGGTGGCATAACCACTTGAATCCA
GAAGTTAAGAAAACCTCCTGGACAGAAGAGGAAGACAGAATTATTTACCAGGCACACAAGAGACTGGGGA
ACAGATGGGCAGAAATCGCAAAGCTACTGCCTGGACGAACTGATAATGCTATCAAGAACCACTGGAATTC
TACAATGCGTCGGAAGGTCGAACAGGAAGGTTATCTGCAGGAGTCTTCAAAAGCCAGCCAGCCAGCAGTG
GCCACAAGCTTCCAGAAGAACAGTCATTTGATGGGTTTTGCTCAGGCTCCGCCTACAGCTCAACTCCCTG
CCACTGGCCAGCCCACTGTTAACAACGACTATTCCTATTACCACATTTCTGAAGCACAAAATGTCTCCAG
TCATGTTCCATACCCTGTAGCGTTACATGTAAATATAGTCAATGTCCCTCAGCCAGCTGCCGCAGCCATT
CAGAGACACTATAATGATGAAGACCCTGAGAAGGAAAAGCGAATAAAGGAATTAGAATTGCTCCTAATGT
CAACCGAGAATGAGCTAAAAGGACAGCAGGTGCTACCAACACAGAACCACACATGCAGCTACCCCGGGTG
GCACAGCACCACCATTGCCGACCACACCAGACCTCATGGAGACAGTGCACCTGTTTCCTGTTTGGGAGAA
CACCACTCCACTCCATCTCTGCCAGCGGATCCTGGCTCCCTACCTGAAGAAAGCGCCTCGCCAGCAAGGT
GCATGATCGTCCACCAGGGCACCATTCTGGATAATGTTAAGAACCTCTTAGAATTTGCAGAAACACTCCA
ATTTATAGATTCTGATTCTTCATCATGGTGTGATCTCAGCAGTTTTGAATTCTTTGAAGAAGCAGATTTT
TCACCTAGCCAACATCACACAGGCAAAGCCCTACAGCTTCAGCAAAGAGAGGGCAATGGGACTAAACCTG
CAGGAGAACCTAGCCCAAGGGTGAACAAACGTATGTTGAGTGAGAGTTCACTTGACCCACCCAAGGTCTT
ACCTCCTGCAAGGCACAGCACAATTCCACTGGTCATCCTTCGAAAAAAACGGGGCCAGGCCAGCCCCTTA
GCCACTGGAGACTGTAGCTCCTTCATATTTGCTGACGTCAGCAGTTCAACTCCCAAGCGTTCCCCTGTCA
AAAGCCTACCCTTCTCTCCCTCGCAGTTCTTAAACACTTCCAGTAACCATGAAAACTCAGACTTGGAAAT
GCCTTCTTTAACTTCCACCCCCCTCATTGGTCACAAATTGACTGTTACAACACCATTTCATAGAGACCAG
ACTGTGAAAACTCAAAAGGAAAATACTGTTTTTAGAACCCCAGCTATCAAAAGGTCAATCTTAGAAAGCT
CTCCAAGAACTCCTACACCATTCAAACATGCACTTGCAGCTCAAGAAATTAAATACGGTCCCCTGAAGAT
GCTACCTCAGACACCCTCTCATCTAGTAGAAGATCTGCAGGATGTGATCAAACAGGAATCTGATGAATCT
GGAATTGTTGCTGAGTTTCAAGAAAATGGACCACCCTTACTGAAGAAAATCAAACAAGAGGTGGAATCTC
CAACTGATAAATCAGGAAACTTCTTCTGCTCACACCACTGGGAAGGGGACAGTCTGAATACCCAACTGTT
CACGCAGACCTCGCCTGTGGCAGATGCACCGAATATTCTTACAAGCTCCGTTTTAATGGCACCAGCATCA
GAAGATGAAGACAATGTTCTCAAAGCATTTACAGTACCTAAAAACAGGTCCCTGGCGAGCCCCTTGCAGC
CTTGTAGCAGTACCTGGGAACCTGCATCCTGTGGAAAGATGGAGGAGCAGATGACATCTTCCAGTCAAGC
TCGTAAATACGTGAATGCATTCTCAGCCCGGACGCTGGTCATGTGA
>NM_001130172.1
ATGGCCCGAAGACCCCGGCACAGCATATATAGCAGTGACGAGGATGATGAGGACTTTGAGATGTGTGACC
ATGACTATGATGGGCTGCTTCCCAAGTCTGGAAAGCGTCACTTGGGGAAAACAAGGTGGACCCGGGAAGA
GGATGAAAAACTGAAGAAGCTGGTGGAACAGAATGGAACAGATGACTGGAAAGTTATTGCCAATTATCTC
CCGAATCGAACAGATGTGCAGTGCCAGCACCGATGGCAGAAAGTACTAAACCCTGAGCTCATCAAGGGTC
CTTGGACCAAAGAAGAAGATCAGAGAGTGATAGAGCTTGTACAGAAATACGGTCCGAAACGTTGGTCTGT
TATTGCCAAGCACTTAAAGGGGAGAATTGGAAAACAATGTAGGGAGAGGTGGCATAACCACTTGAATCCA
GAAGTTAAGAAAACCTCCTGGACAGAAGAGGAAGACAGAATTATTTACCAGGCACACAAGAGACTGGGGA
ACAGATGGGCAGAAATCGCAAAGCTACTGCCTGGACGAACTGATAATGCTATCAAGAACCACTGGAATTC
TACAATGCGTCGGAAGGTCGAACAGGAAGGTTATCTGCAGGAGTCTTCAAAAGCCAGCCAGCCAGCAGTG
GCCACAAGCTTCCAGAAGAACAGTCATTTGATGGGTTTTGCTCAGGCTCCGCCTACAGCTCAACTCCCTG
CCACTGGCCAGCCCACTGTTAACAACGACTATTCCTATTACCACATTTCTGAAGCACAAAATGTCTCCAG
TCATGTTCCATACCCTGTAGCGTTACATGTAAATATAGTCAATGTCCCTCAGCCAGCTGCCGCAGCCATT
CAGAGACACTATAATGATGAAGACCCTGAGAAGGAAAAGCGAATAAAGGAATTAGAATTGCTCCTAATGT
CAACCGAGAATGAGCTAAAAGGACAGCAGGTGCTACCAACACAGAACCACACATGCAGCTACCCCGGGTG
GCACAGCACCACCATTGCCGACCACACCAGACCTCATGGAGACAGTGCACCTGTTTCCTGTTTGGGAGAA
CACCACTCCACTCCATCTCTGCCAGCGGATCCTGGCTCCCTACCTGAAGAAAGCGCCTCGCCAGCAAGGT
GCATGATCGTCCACCAGGGCACCATTCTGGATAATGTTAAGAACCTCTTAGAATTTGCAGAAACACTCCA
ATTTATAGATTCTTTCTTAAACACTTCCAGTAACCATGAAAACTCAGACTTGGAAATGCCTTCTTTAACT
TCCACCCCCCTCATTGGTCACAAATTGACTGTTACAACACCATTTCATAGAGACCAGACTGTGAAAACTC
AAAAGGAAAATACTGTTTTTAGAACCCCAGCTATCAAAAGGTCAATCTTAGAAAGCTCTCCAAGAACTCC
TACACCATTCAAACATGCACTTGCAGCTCAAGAAATTAAATACGGTCCCCTGAAGATGCTACCTCAGACA
CCCTCTCATCTAGTAGAAGATCTGCAGGATGTGATCAAACAGGAATCTGATGAATCTGGAATTGTTGCTG
AGTTTCAAGAAAATGGACCACCCTTACTGAAGAAAATCAAACAAGAGGTGGAATCTCCAACTGATAAATC
AGGAAACTTCTTCTGCTCACACCACTGGGAAGGGGACAGTCTGAATACCCAACTGTTCACGCAGACCTCG
CCTGTGGCAGATGCACCGAATATTCTTACAAGCTCCGTTTTAATGGCACCAGCATCAGAAGATGAAGACA
ATGTTCTCAAAGCATTTACAGTACCTAAAAACAGGTCCCTGGCGAGCCCCTTGCAGCCTTGTAGCAGTAC
CTGGGAACCTGCATCCTGTGGAAAGATGGAGGAGCAGATGACATCTTCCAGTCAAGCTCGTAAATACGTG
AATGCATTCTCAGCCCGGACGCTGGTCATGTGA
Muscle
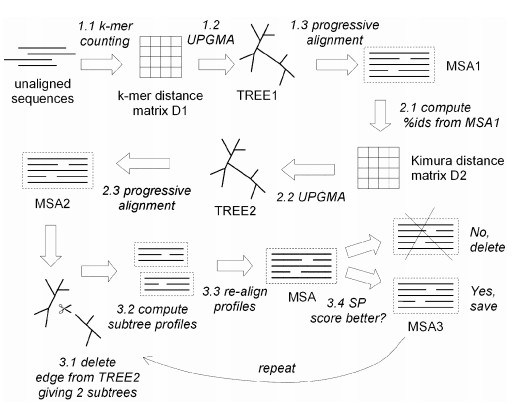 Iterative Progressive Alignment
Iterative Progressive Alignment
muscle --help
muscle -in example.aa -out example.aa.muscle
muscle -in example.aa -out example.aa.muscle.clw -clw
muscle -in example.fasta -out example.fasta.muscle
muscle -in example.fasta -out example.fasta.muscle.clw -clw
ClustalO
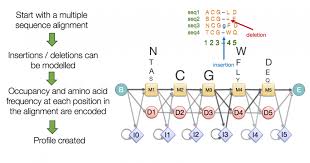 Hidden Markov Model
Hidden Markov Model

https://www.ebi.ac.uk/Tools/msa/clustalo/
clustalo --help
clustalo -i example.aa -o example.aa.clustao
clustalo -i example.aa -o example.aa.clustao.clw --outfmt=clu
clustalo -i example.fasta -o example.fasta.clustalo
clustalo -i example.fasta -o example.fasta.clustalo.clw --outfmt=clu
Alignmnet Visualization
https://www.ebi.ac.uk/Tools/msa/mview/