Installing Packages Using Conda
Conda is a package manager, which helps you find and install packages such as numpy or scipy. It also serves as an environment manager, and allows you to have multiple isolated environments for different projects on a single machine. Each environment has its own installation directories, that doesn’t share packages with other environments.
For example, you need python 2.7 and Biopython 1.60 in project A, while you also work on another project B, which needs python 3.5 and Biopython 1.68. You can use conda to create two separate environments for each project, and you can switch between different versions of packages easily to run your project code.
Install Python package without Conda
Pip, which stands for Pip Install Packages, is Python’s official package manager. We can install packages through pip. You can find the list of available packages from Python Package Index (PyPI) https://pypi.python.org/pypi You already have pip, if your python 2 >= 2.7.9 or Python 3 >= 3.4. Otherwise you need to install pip, according to the instructions. In terminal, you enter the following to install a package.
First, make sure that your package is in the pip:
pip search <package>Then, you can install this package:
pip install <package>
Why conda?
-
Dependencies is one of the main reasons to use Conda. Sometimes, install a package is not as straight forward as you think. Imagine a case like this: You want to install package Matplotlib, when installing, it asks you to install Numpy, and Scipy, because Matplotlib need these Numpy and Scipy to work. They are called the dependencies of Matplotlib. For Numpy and Scipy, they may have their own dependencies. These require even more packages.
-
Conda provide a solution for this situation: when you install package Matplotlib, it will automatically install all the dependencies like Numpy and Scipy. So you don’t have to install them one by one, manually. This can save you great amount of time.
-
The other advantage of conda, is that conda can have multiple environments for different projects. As mentioned at the very beginning, it can have two separate environments of different versions of software. Using conda environment on BioHPC
Check python
First, you need to check python Version 2.7.x Or version 3.5.x.
python -v
If you don’t know how to. Please look at other pages Python version control
Python 3 is the latest version of the language and python 2 is legacy. So you should choose Python 3.7.x when you could. However, if you know some packages you want are not compatible with Python 3, then you can install Python 2.7.x. See here for details about Python 2 and Python3.
Anaconda or Miniconda?
-
Anaconda includes both Python and conda, and additionally bundles a suite of other pre-installed packages geared toward scientific computing. Because of the size of this bundle, expect the installation to consume several gigabytes of disk space.
-
Miniconda gives you the Python interpreter itself, along with a command-line tool called conda which operates as a cross-platform package manager geared toward Python packages, similar in spirit to the apt or yum tools that Linux users might be familiar with.
Install Miniconda
Visit the miniconda page and get ready to download the installer of your choice/system.
For linux:
$ wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
For Mac:
$ wget https://repo.anaconda.com/miniconda/Miniconda3-latest-MacOSX-x86_64.sh
Example for linux:
$ chmod +x Miniconda-latest-Linux-x86_64.sh
$ ./Miniconda-latest-Linux-x86_64.sh
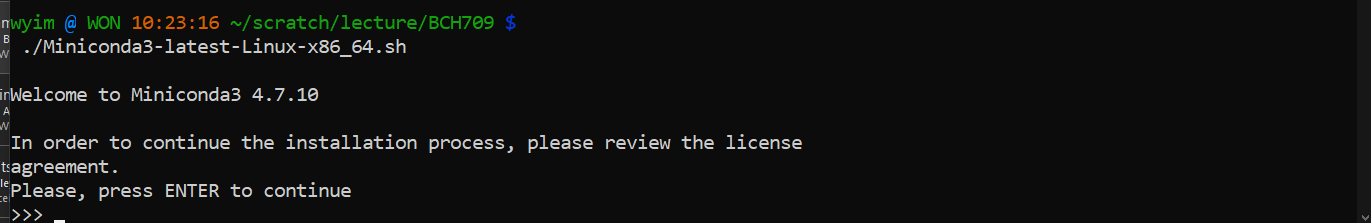

Reload your enviroment
$ source ~/.bashrc
Initialize Miniconda3
$ conda init
Create new conda environment
Create a conda environment named test with latest anaconda package.
$ conda create -n bch709 python=3
Alternatively you can specify python version
$ conda create -n snowdeer_env python=2.7.16
*Usually, the conda environment is installed in your home directory on computer, /home/<your username>. The newly created environment
Use the environment you just created
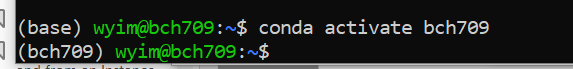
Activate your environment:
$ conda activate bch709
It will show your environment name at the beginning of the prompt.
Install packages in the conda environment
Install from default conda channel You can search if your package is in the default source from Anaconda collection. Besides the 200 pre-built Anaconda packages, it contains over 600 extra scientific and analytic packages. All the dependencies will also be installed automatically.
$ conda search <package>
$ conda install <package>
Install from conda-forge channel (example: hisat2)
Conda channels are the remote repository that conda takes to search or download the packages. If you want to install a package that is not in the default Anaconda channel, you can tell conda which channel containing the package, so that conda can find and install. Conda-forge is a GitHub community-led conda channel, containing general packages which are not in the default Anaconda source. All the packages from conda-forge is listed at https://bioconda.github.io/conda-recipe_index.html
$ conda search hisat2
$ conda search -c bioconda hisat2
Install from bioconda channel (example: stringtie)
Bioconda is another channel of conda, focusing on bioinformatics software. Instead of adding “-c” to search a channel only one time, “add channels” tells Conda to always search in this channel, so you don’t need to specify the channel every time. Remember to add channel in this order, so that Bioconda channel has the highest priority. Channel orders will be explained in next part.
$ conda config --add channels conda-forge
$ conda config --add channels defaults
$ conda config --add channels r
$ conda config --add channels bioconda
Adding channels will not generate any command line output. Then, you can install Stringtie from the Bioconda channel
$ conda install hisat2
All the bioconda packages can be found here: https://bioconda.github.io/conda-recipe_index.html
Channel order
If you add multiple channels in one environment using (conda config –add channels
For example, if you add the channels in different order in the Stringtie example, by switching channel r and channel bioconda. Say, channel R has package A version 0.8 and bioconda has A version 1.0. The environment will have A 0.8 now from channel R, since it’s the highest priority. Then, Stringtie might not work if it need package A 1.0.
If the packages can be found in different channels, the package from the highest priority channel will be installed even if the version of it isn’t newest. The command <conda config –append channels new_channel> puts the new channel at the bottom of the channel list, making it the lowest priority.
Install R and R packages
The Conda package manager is not limited to Python. R and R packages are well supported by a conda channel maintained by the developers of Conda. The R-essentials include all the popular R packages with all of their dependencies. The command below opens R channel by “-c r”, and then install the r-essentials using R channel.
$ conda install -c r r-essentials
Update R packages
$ conda update -c r r-essentials
$ conda update -c r r-<package name>
More conda commands:
See all available environments
You can check the list of all separate environments, and it will show * at your current environment. In the figure below, it shows root, since I’m not in any conda environment.
$ conda env list
List all package installed
This will show all the packages and versions you’ve installed.
$ conda list
Update packages or conda itself
This will update to the newest version of one package, or conda itself. update package
$ conda update <package>
update package in env
conda update --name <ENV_name> <package>
update conda itself
$ conda update -n test --all
$ conda update -n bch709 --all
Uninstall package from the environment
$ conda uninstall <package name>
Exit current environment:
You can exit, when you finish your work in the current environment.
$ conda deactivate
Remove environment
$ conda env remove --name bch709
When you finish your project, you might want to remove the environment. However, it is not recommended because you might want to update some work in this project in the future.
Enviroment export
conda env export --name <ENVIRONMENT> --file <outputfilename>.yaml
Envrioment import
conda env create --file <outputfilename>.yaml
Reference:
- Conda documentation https://docs.conda.io/en/latest/
- Conda-forge https://conda-forge.github.io/
- BioConda https://bioconda.github.io/
